Abstract
Influenza viruses are a major public health threat worldwide. The influenza hemagglutinin (HA) plays an essential role in the virus life cycle. Due to the high conservation of the HA stem region, it has become an especially attractive target for inhibitors for therapeutics. In this study, molecular simulation was applied to study the mechanism of a small molecule inhibitor (MBX2329) of influenza HA. Behaviors of the small molecule under neutral and acidic conditions were investigated, and an interesting dynamic binding mechanism was found. The results suggested that the binding of the inhibitor with HA under neutral conditions facilitates only its intake, while it interacts with HA under acidic conditions using a different mechanism at a new binding site. After a series of experiments, we believe that binding of the inhibitor can prevent the release of HA1 from HA2, further maintaining the rigidity of the HA2 loop and stabilizing the distance between the long helix and short helices. The investigated residues in the new binding site show high conservation, implying that the new binding pocket has the potential to be an effective drug target. The results of this study will provide a theoretical basis for the mechanism of new influenza virus inhibitors.
Similar content being viewed by others
Introduction
Influenza virus is the causative agent of influenza, an infectious disease which usually leads to symptoms such as high fever, cough, headache, muscle and joint pain, sore throat, nasal discharge, and even a fatal illness similar to pneumonia1,2,3,4. Influenza viruses are divided into three types, type A, type B and type C, with influenza A virus presenting serious threats to public health worldwide due to its high mutation rate5,6,7. At present, two classes of drugs are approved by the Food and Drug Administration for treatment or chemoprophylaxis of influenza: matrix protein 2 (M2) inhibitors amantadine and rimantadine and the neuraminidase (NA) inhibitors (NAIs) oseltamivir and zanamivir8, 9. However, with the wide use of these drugs, drug-resistant strains have appeared in succession10. Therefore, new antiviral targets with novel inhibition mechanism need to be developed.
Hemagglutinin (HA), a viral receptor-binding and membrane-fusion glycoprotein involved in the invasion of influenza into host cells, plays an essential role in the life cycle of influenza A virus3, 11. HA is a trimer of identical subunits, each of which consists of a variable membrane-distal receptor-binding globular domain (HA1) and a more conserved membrane-proximal helix-rich stem structure (HA2)12. Under acidic conditions, the residues on the outer surface of HA1 can be protonated easily, which leads to the large gathering of positive charges on the surface of HA13, 14. With gradually increased charges, disaggregation of HA first occurs as the positive charges repel each other, followed by the entry of water into the interior of the protein causing further structural changes of HA213. In the HA2 subunit, one short and one long α-helical segment are connected by a loop (helix-loop-helix structure)3, 14, 15. The N-terminal segment next to the short helix is a fusion peptide consisting of 20 amino acids, while the C-terminal end of the long helix is anchored to the viral membrane3. Acidic pH conditions in the endosome can trigger conformational changes involving a folding of the loop connecting the two helices of HA2 into a new longer helix (coiled-coil structure) of the HA ectodomain (loop-to-helix transition) and further trigger its membrane fusion capacity1, 3. From the above observations, stabilizing the “helix-loop-helix” structure of HA2 may be considered crucial for preventing the loop-to-helix transition11, 15.
The sequence and structure of the HA stem loop region are well known to be highly conserved, which makes it an especially attractive target for entry inhibitors for therapeutics16. Currently, many anti-flu drug target studies have focused on this functional membrane protein. For instance, Arnab and colleagues successfully obtained two novel promising compounds, MBX2329 and MBX2546, using a high-throughput screening assay (HTS), a way to screen novel compounds from a chemical library for inhibitory activity of some functional protein, and demonstrated their potency in the inhibition of HA16, 17. Other HA-inhibiting compounds such as BMY 2770918, 180299 (podocarpic acid derivative)19 and tert-butyl hydroquinone (TBHQ)20 have also been confirmed. These compounds can serve as starting points for the development of a therapeutic agent. Therefore, studying the inhibition mechanism of novel compounds in theory is necessary.
It has been confirmed that MBX232916, the compound mentioned above, could inhibit HA of A/Washington/10/2008(H1N1) and A/Florida/21/2008(H1N1) at similar levels (IC50 of 0.29 µM and 0.3 µM). However, it’s still not clear about the theoretical basis of its inhibition mechanism. In order to explore molecular inhibition mechanism of new HA inhibitors, in this study, molecular docking, molecular dynamics simulation and the Molecular Mechanics/Poisson-Boltzmann Surface Area (MM-PBSA) approach were applied to study the detailed mechanism of HA inhibition of this representative compound MBX2329, which hereafter is referred to as INT for convenience16. The findings of this study will be useful for future exploration of efficient drug targets and provide theoretical insight into a new mechanism of influenza virus inhibitors.
Theory and Methods
Preparation of initial complexes
HAs from experimental strains A/Washington/10/2008(H1N1) (HAWashington) and A/Florida/21/2008(H1N1) (HAFlorida)16 were chosen for the preferred models to investigate the INT binding pose under neutral condition, and eventually, the results between experiment and simulation were compared. According to the high homology between HAWashington and HAFlorida, the structure of HAFlorida could be regarded as a repeated system in the following molecular simulations to ensure the accuracy of molecular dynamics sampling. The three-dimensional structures of the above HA were obtained via Swiss-modeling21, 22, a homology modeling procedure, since their structures have not been crystallized. The HA structure from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB ID: 4LXV23) was chosen as the template for homology modeling as its highly BLAST sequence alignment similarity (higher than 87%) to HAWashington’s or HAFlorida’s (Fig. S1a). Procheck24, 25 was used to validate the rationality of homology models built with the Swiss model online. In order to perform a further investigation on inhibition mechanism under acidic condition, a third HA structure of strains 1918(H1N1) (RCSB PDB ID: 1RUZ26) was chosen for the model at a low-pH state according to the calculated pKa results for its titratable residues as studied by Zhou et al.27. Protonation state of the titrable residues in 1RUZ is shown in Table S1. Energy minimizations were performed on all HA with GROMACS 4.5.228 using steepest descent techniques before docking calculations. The structure of INT was optimized at the B3LYP 6–31 + G* level using the Gaussian 09 software29. Relevant structures are shown in Supplementary Materials Fig. S2.
AutoDock Vina30 was used for automatic placement of INT in the binding pockets of HAs to obtain the initial structure of INT-HA complexes. AutoDock Vina is a new program for molecular docking and virtual screening, which uses a sophisticated gradient optimization method in its local optimization procedure. For AutoDock Vina, a grid of 24 × 22 × 24 points in the x, y, and z-axis directions was built with a grid spacing of 1 Å, the center of which was treated as the mass center of one monomer of HA, and the exhaustiveness was set to 2031. The grid detail used in the docking simulations is shown in Fig. S3.
Molecular dynamics simulation
The three molecular systems are listed in Table 1. All of the complex systems were subjected to molecular dynamics simulation in a periodic boundary condition using the GROMACS 4.5.2 software package28 with SPC (simple point charge) water model32. The Gromos 53 A6 force field33 was applied to describe both the receptors and ligand. The parameterization of the INT was produced by Automated Topology Builder (ATB) server34. To keep each system at an electrically neutral state, an appropriate amount of ions was added to randomly replace the water molecules (18 sodium ions for INT-HAWashington complex, 15 sodium ions for INT-HAFlorida complex and 33 chloride ions for INT-HA1ruz complex). First, energies of the complex systems were relaxed with steepest-descent energy minimization to ensure that the systems were without steric clashes or incorrect geometry. Thereafter, 500 ps NVT (constant Number of particles, Volume, and Temperature) and NPT (constant Number of particles, Pressure, and Temperature) were alternately run with position restraints on HA and INT to allow for relaxation of the solvent molecules in two phases. The solvent molecules were equilibrated with fixed protein at 300 K, taking the initial velocities from a Maxwellian distribution. Subsequently, the protein and inhibitor were relaxed step by step and heated up to 300 K. The long-range electrostatics were described with the particle mesh Ewald algorithm35 with interpolation order of 4, a grid spacing of 0.16 nm and the Coulomb cutoff distance of 1.0 nm. The LINCS (Linear Constraint Solver) algorithm was used to constrain all bonds. Temperature and pressure coupling types were set with V-rescale and Parrinello-Rahman, respectively31, 36. In the NVT ensemble, the temperature of the systems reached a plateau at the desired value (reference temperature = 300 K; time constant = 0.1 ps). In addition, the equilibration of pressure (reference pressure = 1.0 bar; time constant = 2.0 ps) was performed under the NPT ensemble. Finally, molecular dynamics simulations for collecting data with a time step of 2 fs and coordinates saved every 2 ps were initiated29, 31.
Calculation of binding free energy and ligand-residue interaction decomposition
In our study, the binding free energies were calculated using the MM-PBSA method37,38,39 supplied with the Amber 10 package40. A total number of 100 snapshots were chosen evenly from the molecular dynamics trajectory31. The MM-PBSA method can be conceptually summarized as:
where the ΔH of the system consists of the enthalpy changes in the gas phase upon complex formation (ΔE MM ) and the solvated free energy contribution (ΔG sol ), while −TΔS refers to the entropy contribution to the binding31. Equation (2) can then be approximated as shown in Eq. (3):
In addition, the change in flexibility of HA and inhibitor upon binding is similar in all complexes. Based on previous studies41, the entropy differences should be very small. Thus, the calculation of the solute entropy term was omitted in the present study31, 42.
ΔE MM is the summation of the van der Waals (ΔE vdw ) and the electrostatic (ΔE ele ) interaction energies.
In addition, ΔG sol , which denotes the solvation free energy, can be computed as the summation of an electrostatic component (ΔG ele,sol ) and a nonpolar component (ΔG nonpolar,sol ), as shown in Eq. (5). The nonpolar contribution to the free energy was calculated via γSASA, where SASA is the solvent-accessible surface area and γ is 0.0072 kcal/mol/Å2. The SASA was estimated to be a 1.4 Å solvent-probe radius. Referring to nonpolar solvation options the radii used in the MM-PBSA calculations are mbondi2.
The interactions between the inhibitor and each residue in the binding site were analyzed using the MM-PBSA decomposition process applied in the MM-PBSA module in Amber 10. The binding interaction of each ligand-residue pair includes three terms: the van der Waals contribution (ΔE vdw ), the electrostatic contribution (ΔE ele ) and the solvation contribution (ΔE sol ). All energy components were calculated using the same snapshots as the free energy calculation31, 42.
Results and Discussion
Influenza virions that are attached to the host cell membrane via HA are phagocytosed and gain entry into the cell through endocytosis11. During this endocytosis process, small molecules generally can enter the endosome at the same time43. Thus, the effective combination of Inhibitors of HA under neutral conditions outside the cell is crucial for the intake of INT. After entering into the endosome, HA will undergo large conformational changes under acidic conditions, which is aimed at promoting the membrane fusion between the virus and host cell1, 3. An efficient HA entry inhibitor must be able to stop the progression of changes in this molecule. In order to investigate if INT could stably bind with HA under neutral conditions and be efficiently carried into the endosome to exert its inhibition at low-pH conditions, the optimal binding mode, binding force and binding behavior of INT with HA were determined by a series of molecular dynamics simulations and MM-PBSA calculation.
Analysis of reliability of investigated complex systems
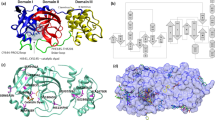
The validation was carried out using Ramachandran plot calculations computed with the Procheck24, 25 program by examining the detailed residue-by-residue stereochemical quality of HA structures, and the results are shown in Fig. 1. Altogether, 100% of the investigated residues were in allowed regions, which confirmed the availability of the modeled HA protein systems. AutoDock Vina was used for obtaining the initial structure of INT-HA complexes. The docking results were in agreement with those from the mutation experiments and molecular docking reported by Arnab et al.17, which indicated that the findings could be applied to the next molecular dynamics study. Relevant docking results (Fig. S4) and comparisons with previous results from Arnab et al. can be found in the Supplementary Materials.
For the accuracy of dynamics sampling, the complex structure of INT-HAFlorida was regarded as a replicate system of INT-HAWashington under the neutral condition molecular simulations. In order to monitor if two systems possess similar dynamics behaviors, the cross-correlations of their Cα atomic displacements, a parameter which can reflect the detailed atomic dynamic state, were analyzed during the simulations29. The relevant results are illustrated in Fig. 2. Highly positive regions (red) indicate a strong correlation in the movement of residues, whereas negative regions (blue) are associated with strong anti-correlated motion of the residues. The color depth of diagonal could represent the movement degree of atoms. As shown in Fig. 2a and b, the similarity could be observed in the behavior of Cα atoms from HAWashington HA1 and HAFlorida HA1. Also the similar behavior could be discovered from Cα atoms from HAWashington HA2 and HAFlorida HA2 as shown in Fig. 2c and d. As expected, no obvious differences in the movement of atoms occurred between the two complexes during the timescale of simulations. Moreover, according to the structures of both simulations, the conformations of two repeated simulations were found to be identical. Two independent dynamics samplings yielded similar results. Thus, in the following sections, results from only INT-HAWashington are discussed.
Cross-correlation matrix of the fluctuations of each of the x, y, and z coordinates of the Cα atoms of HA from their average during molecular dynamics simulations. (a) Cα atoms from HA1 of HAWashington. (b) Cα atoms from HA1 of HAFlorida. (c) Cα atoms from HA2 of HAWashington. (d) Cα atoms from HA2 of HAFlorida.
Binding mode of complex system under neutral conditions
For convenience, INT-HAWashington is simplified as INT-HA in the following section. After 200 ns simulations, the root-mean-square deviation (RMSD) of backbone Cα atoms and all atoms of HA were first investigated to evaluate if the complex system could reach an equilibrium during the simulation44. As shown in Fig. 3, the RMSD curve of the INT-HA complex was found to reach the equilibrium at about 40 ns, suggesting that the structures could be applied to analyze the optimal binding mode between INT and HA31.
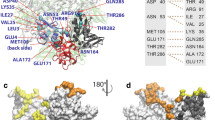
In order to obtain the most stable complex structure, cluster analysis of INT-HA was investigated to determine the optimal binding mode. In addition, the comparison of free energy surfaces between the first 100 ns and further one with extended 100 ns were applied to confirm the convergence of complexes (Fig. S5). Cluster analysis is a method that can categorize similar low-energy conformations into the same designated area by analyzing the relative free energy surfaces along the first two principle components (PC-1, PC-2) of the complex during simulation29, 33. Two principal amounts (PC-1 and PC-2) were obtained as the reaction coordinate that renders the potential of the mean force (ΔF = −RT × lnP, where P is the relative probability in a region) free-energy plane42. The first few PCs describe the slow-motion modes which are related to the functional motions of a biomolecular system. In the cluster analysis plot (Fig. 4a), the conformations found in the blue area indicated more stable and lower energy states than those found in the red area. And these lower energy conformations generally could be chosen as the best analysis subjects for the binding modes. As shown in Fig. 4a, more than 80% of the conformations converged at the blue area, and the structure from this area could be applied to the binding mode analysis. The detailed binding mode of INT-HA based on the above analysis is shown in Fig. 4b. The results revealed that INT is bound in the pocket located on the interface between HA1 and HA2. This pocket is surrounded by the side chains of HA1-Val31, HA1-Leu290, HA1-Thr316, HA2-Ile47, HA2-Thr48 and HA2-Val51. The side chain of HA1-Val31 extends towards C9 of INT, and the side chain of HA1-Leu290 is close to C7 of INT. HA1-Thr316, HA2-Ile47, HA2-Thr48 and HA2-Val51 together approach the seven-member heterocycle of INT.
In order to confirm if INT could stably bind with HA in this pocket during the simulation, the distances between the above residues and INT were calculated by using the distance analysis program of GROMACS. According to the corresponding results shown in Fig. 5, all of the distances monitored were around 0.3–0.4 nm during the 200 ns simulation, which indicated the accuracy of the binding pose. INT is a molecule with an aminoalkyl phenol ether scaffold16 and has no hydrogen donor or receptor. Thus, the stabilization of INT in the binding pocket most likely contributes to nonpolar interactions. It is well known that the van der Waals radius of a carbon atom is 0.2 nm, and distances of 0.3–0.4 nm favor the formation of van der Waals interactions between two carbon atoms. According to the distances between INT and residues around the pocket shown in Fig. 5, we speculate that van der Waals interactions provide the major contribution to the bonding of INT and HA. This speculation was verified as described in the next section.
Verification of binding mode of INT-HA under neutral conditions
In order to further validate the binding modes and obtain detailed information on the key residues of the binding pocket in the complex, the electrostatic, van der Waals, solvation and total contribution of the residues to the binding free energy of the INT-HA complex were calculated with the MM-PBSA method. To achieve more accurate results, MM-PBSA calculations were carried out twice. The snaps used in calculations were respectively extracted from equilibrium stages within first and second 100 ns. By comparing the energy of the two results, similarity could be observed (Fig. S6), thus, only one group of data from the equilibrium stages within the first 100 ns was shown in this section. The contributions from the residues around INT are shown in Fig. 6 and Table 2.
The contribution of selected residues surrounding the INT-HA complex binding pocket can be clearly seen from Fig. 6 and Table 2. As shown, HA1-Val31, HA1-Leu290, HA1-Thr316, HA2-Ile47, HA2-Thr48 and HA2-Val51 have favorable total energy contributions, suggesting that these residues participate in the complex binding. More importantly, for aforementioned residues van der Waals interactions play a major role in the total energy contribution as evidenced by lower values of ΔE vdw (−1.72, −0.65, −0.52, −0.54, −0.73 and −1.08 kcal·mol−1, respectively). The free energy calculation results are consistent with the speculation above that van der Waals interactions are a major contributor to the bonding of INT and HA.
The total binding energy of the INT-HA complex systems, an important standard measure of binding affinity between INT and HA, were calculated as well. Total binding energies of INT-HAWashington and INT-HAFlorida were calculated by MM-PBSA at the same time. As listed in Table 3, the results show that the values of ΔG total for INT-HAWashington and INT-HAFlorida are −16.15 and −16.02 kcal·mol−1, respectively, indicating that INT has similar binding interactions with HAWashington and HAFlorida. The calculated result is qualitatively consistent with the experimental results showing that INT could inhibit HA of A/Washington/10/2008(H1N1) and A/Florida/21/2008(H1N1) at similar levels (IC50 of 0.29 µM and 0.3 µM) in vitro 16.
Contributions from total free energy were analyzed in detail as well. The ΔG total can be divided into polar and nonpolar energies. The free energies of INT binding to two HAs were found to be primarily derived from the nonpolar energies (equal to ΔG np + ΔE vdw ), which were −21.95 kcal·mol−1 in the INT-HAWashington complex and −22.75 kcal·mol−1 in the INT-HAFlorida complex. However, for both complexes, the polar energy (equal to ΔG PB + ΔE ele ) showed negative contributions at 5.80 and 6.73 kcal·mol−1 for INT-HAWashington and INT-HAFlorida, respectively. In addition, the values of ΔE vdw for INT-HAWashington and INT-HAFlorida were calculated at −18.29 and −19.42 kcal·mol−1, respectively, indicating that van der Waals interactions are dominant in the total energy contribution. The total free energy calculations also were in agreement with the speculation above, indicating that nonpolar interactions, especially van der Waals interactions, are the major contributor to the bonding of INT and HA.
Thus, the above analysis suggests that INT can stably bind with HA under neutral conditions via van der Waals interactions and has the ability to enter the endosome following HA binding during endocytosis.
Binding and dynamics behavior of INT at low pH conditions
In the above discussion, effective binding of INT with HA under neutral conditions was demonstrated. Nevertheless, the ability of INT to maintain the interaction with HA at a low pH is crucial to its inhibition and determines whether it has the potential to be a HA entry inhibitor. Prior experimental data indicated that INT indeed could inhibit different influenza virus H1N1 strains in vitro 16, but its mechanism of inhibition at low pH at the atom level has not been explored. In this section, the binding and dynamics behavior of INT at low-pH conditions will be discussed.
As described in method, HA1ruz was chosen as the low-pH model in this section based on the calculated pKa of the titratable residues by Zhou et al. In order to determine whether the INT binding would affect the protonation state of the residues, the titratable residues’ locations of HA1ruz were investigated. Most titratable residues were located far away from the INT binding site except HA1-Lys278, HA1-Glu302 and HA2-Glu56. Subsequently, the pKa of the above three residues were calculated via PROPKA45, 46 with or without INT (Table S2). It’s further illustrated that the bound of INT could not affect the protonation state of the residues. Thus, protonation state of HA1ruz could be assigned according to the results of Zhou et al. directly (Table S1).
In order to facilitate the comparison, for the investigated complex in this study only one monomer of the HA trimer was bound with INT, and the other two unbound monomers still could disaggregate at low pH as usual. In order to monitor the location of INT at low-pH conditions, distances between INT and its binding residues (HA1-Val31, HA1-Leu290, HA1-Thr316, HA2-Ile47, HA2-Thr48 and HA2-Val51) in the neutral state were calculated again. As shown in Fig. 7, almost all the distances between INT and its binding residues (in neutral state) were disrupted at different degrees at 0–45 ns and increased sharply at about 46 ns, except for HA1-Leu290. The disrupted distances kept at about 65–200 ns with small fluctuation. These observations suggest that INT had relocated out of the pocket formed by the side chain of HA1-Val31, HA1-Leu290, HA1-Thr316, HA2-Ile47, HA2-Thr48 and HA2-Val51 and moved to a new low potential energy point at 65–200 ns. The distance between INT and Leu290-HA1 was always maintained at about 0.3–0.4 nm, which indicated that the new binding location was still nearby. In order to confirm this phenomenon, an independent repeated trial was conducted with a different computing resource with the same parameters, and the same results were obtained (Fig. S7). Therefore, we suspected that the binding of INT with HA under neutral conditions only facilitates its intake, and another mechanism exists for the INT binding and inhibition of HA at low-pH conditions.
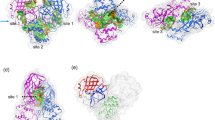
In order to explore the binding and dynamic behavior of INT at low pH, a cluster analysis of INT-HA1ruz was performed. As shown in Fig. 8a, unlike with INT-HA under neutral conditions (Fig. 4a), mainly five blue areas could be found in the free energy surface plot of the complex at low pH. Most conformations of 1–8 ns, 9–28 ns, 29–64 ns, 65–145 ns and 146–200 ns were clustered in regions 1, 2, 3, 4 and 5, respectively. Five representative conformations extracted from different areas are shown in Fig. 8b. After analysis, INT could bind with HA in at least four dominant positions at low-pH conditions. In the initial stage, INT bound between HA1 and the short helix of HA2, but INT moved to the HA head region gradually over time. According to the cluster analysis, INT kept the fourth binding state (Fig. 8b (4 and 5)) with HA after 65 ns, which is in agreement with the results from Fig. 7.
Due to the bulky head region of HA1 being rich in protonated residues at low pH, a large repulsion promoted the structural disaggregation at the initial stage of the simulation13, 14. As the initial binding site of INT was far away from the positively charged head, INT could not prevent the disaggregation at the initial stage. We propose that following the disaggregation of HA1 and the shift of charged state of HA2, a new binding site might be revealed and thus enabled the purposeful relocation of INT46.
The new detailed binding mode of the INT-HA1ruz complex at low pH was investigated and verified by simulation and MM-PBSA calculation twice as described above (Fig. S8). The relevant results are provided in Fig. 9 and Table 4. As shown, a new pocket bound by INT at low pH was found to be surrounded by HA1-Lys278, HA1-Ser289, HA1-Leu290, HA1-Pro291, HA1-Pro304, HA2-Ile55, HA2-Glu56 and HA2-Thr60. In detail, the seven-member heterocycle of INT could locate between side chains of HA1-Lys278 and HA1-Ser289. Side chain of HA1-Pro304 is close to C9 of INT and the side chain of HA2-Thr60 is adjacent to C8 of INT. HA1-Leu290, HA1-Pro291, HA2-Ile55 and HA2-Glu56 together are approaching the six-member ring of INT. The exposed new binding site is still located on the interface between HA1 and HA2. According to the data calculated by MM-PBSA, van der Waals interactions were confirmed to play a major role in the total energy contribution for aforementioned residues as well as evidenced by lower values of ΔE vdw (−1.07, −1.58, −1.72, −2.03, −1.43, −0.58, −1.03 and −0.98 kcal·mol−1, respectively) as shown in Table 4. Although there are four polar residues, HA1-Lys278, HA1-Ser289, HA2-Glu56 and HA2-Thr60, locating around the new binding site, the polarity of these residues still could not contribute to INT binding due to the molecular property of INT. Furthermore, all the polar part of the side chains from the above residues were redirected to the outside of the binding pockets. The sequence conservation of eight residues around the new binding site was evaluated by sequence alignment of the different H1N1 influenza virus strains in the recent 30-year period of time. The results (Table 5) indicated that all the investigated residues in the new binding site are highly conserved, implying that the new binding pocket has the potential to be an effective drug target.
Mechanism of INT Inhibiting HA disaggregation at low pH conditions
Based on analysis of its amino acid sequence, the HA2 polypeptide was found to contain the well-known heptad-repeat module of hydrophobic/hydrophilic amino-acids, which is crucial for the formation of coiled-coil structures. In order to drive the loop-to-helix transition and the formation of a coiled-coil structure, it is necessary for water molecules to directly interact with the hydrophilic residues15. Therefore, the ability of INT to stabilize the connection between HA1 and HA2 and to effectively prevent water from entering into the interior of HA2 is crucial for the inhibitory activity of INT. Because INT is located on the interface between HA1 and HA2, it is considered to not only be able to stabilize HA1 and HA2 but also prevent the entry of water into the interior of HA and further influence the rearrangement of HA2.
In order to investigate the different degrees of water accessibility, the hydrophobic areas of HA2 subunits with or without INT were monitored during 200 ns dynamics simulation at low pH. As shown in Fig. 10, the hydrophobic area of HA2 subunits bound with INT always stabilized at about 70 nm2, but those of the other two free monomers (HA monomers bound without INT) clearly decreased at a value below 70 nm2 as the simulation. The reduced hydrophobic area of the free HA2 subunit indicated that the solution gained access to the inside of the protein due to the lack of protection by INT. By contrast, the HA2 subunit from monomers bound with INT consistently retained the larger hydrophobic area throughout the simulation.
A helix-loop-helix structure provides stability for HA2, and the entry of water must lead to a structural change of this molecule to some extent. We investigated the difference between HA bound with INT and free HAs. As shown in Fig. 11a(1), due to the bound INT, HA preferably maintained the initial helix-loop-helix structure, while the short helixes of the two free HA clearly changed. The secondary structures of the three short helixes were monitored during the 200 ns simulation. The command “do_dssp” of Gromacs was used for secondary structure calculation. As shown in Fig. 11b, the great structural fluctuations in the short helixes of free HAs could be observed at about 30 ns, and the distorted short helixes could endure till 200 ns. These distortions were mainly composed of the formations of the coil, bend and turn during the conformation simulation. By contrast, it’s found that the corresponding secondary structure of HA could display much more stability in the presence of INT. Except for the influence of INT on the short helix, we suspected that the binding of INT also could maintain the stability of the loops connecting the short and long helices. The stability of loops from each HA2 subunit was measured by the RMSD of loops. As shown in Fig. 12a, the RMSD of the loop from the monomer bound with INT was very stable and held the value at about 0.2 nm, indicating that the binding of INT could maintain the rigidity of the loop. RMSD of loops from the two free monomers fluctuated more obviously, which indicated a disruption in the rigidity of the loops. Differences in the loops can be observed in Fig. 11a as well.
In addition, the side chain dynamic behavior of a group of reported residues (Glu101 and Lys50) was also monitored. The salt bridge between Glu101 and Lys50 is crucial for the stabilization of short and long helices of HA2, and the break of this salt bridge would promote the loop-helix transformation27. Therefore, stabilization in the distance between these two residues is an important measure of protective efficacy of INT. From the results shown in Fig. 12b, the distances between Glu101 and Lys50 in the three monomers were found to be different. In the two free monomers, this distance was clearly increased and not favorable for keeping the salt bridge. By contrast, the corresponding distance in the monomer bound with INT was always stable at a lower value, which indicated that the salt bridge of this monomer was protected well and the helix-loop-helix was more stable.
According to the above analysis, we may conclude that INT has the potential to effectively prevent water from entering into the interior of HA, maintain the rigidity of the HA2 loop and stabilize the distance between long helix and short helix, and further obstruct the loop-to-helix transition.
Conclusion
In the current study, the mechanism of binding and inhibition of INT, a potential influenza virus HA inhibitor, was explored by molecular dynamics simulation and MM-PBSA calculation. The behaviors of INT under neutral and acidic conditions were investigated at the same time, and an interesting dynamic binding mechanism of INT was found. We propose that the binding of INT with HA under neutral conditions allows for its intake, while the interaction of INT with HA at low-pH conditions at a new binding site employs a different mechanism to carry out its inhibitory effect. After a series of confirmation, we believe that the binding of INT can prevent HA1 from disassociating from HA2, further maintain the rigidity of the HA2 loop and stabilize the distance between the long helix and short helix.
The investigated residues in the new binding site are highly conserved, implying that the new binding pocket has the potential to be an effective drug target. The results in this study will be useful for the development of efficient drug targets and provide a theoretical basis for the mechanism of new influenza virus inhibitors.
References
Fontana, J. & Steven, A. C. Influenza virus-mediated membrane fusion: Structural insights from electron microscopy. Arch. Biochem. Biophy. 581, 86–97 (2015).
Gong, X. et al. Evaluation of the immunogenicity and protective effects of a trivalent chimeric norovirus P particle immunogen displaying influenza HA2 from subtypes H1, H3 and B. Emerg. Microbes. Infec. 5, e51 (2016).
Lin, X. et al. Order and disorder control the functional rearrangement of influenza hemagglutinin. P. Natl. Acad. Sci. 111, 12049–12054 (2014).
Wu, Y. et al. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci. Rep. 3 (2013).
Otterstrom, J. J. et al. Relating influenza virus membrane fusion kinetics to stoichiometry of neutralizing antibodies at the single-particle level. P. Natl. Acad. Sci. 111, E5143–E5148 (2014).
Gong, X. et al. Conserved stem fragment from H3 influenza hemagglutinin elicits cross-clade neutralizing antibodies through stalk-targeted blocking of conformational change during membrane fusion. Immunol. Lett. 172, 11–20 (2016).
Kanekiyo, M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature. 499, 102–106 (2013).
Ison, M. G. Antivirals and resistance: influenza virus. Curr. Opin. Virol. 1, 563–573 (2011).
Woods, C. J., Malaisree, M., Long, B., McIntosh-Smith, S. & Mulholland, A. J. Computational assay of H7N9 influenza neuraminidase reveals R292K mutation reduces drug binding affinity. Sci. Rep. 3, 3561 (2013).
McKimm-Breschkin, J. L. Influenza neuraminidase inhibitors: antiviral action and mechanisms of resistance. Influenza and other respiratory viruses. 7, 25–36 (2013).
Madhusoodanan, M. & Lazaridis, T. Investigation of pathways for the low-pH conformational transition in influenza hemagglutinin. Biophys. J. 84, 1926–1939 (2003).
Barbey-Martin, C. et al. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 294, 70–74 (2002).
Huang, Q., Opitz, R., Knapp, E.-W. & Herrmann, A. Protonation and stability of the globular domain of influenza virus hemagglutinin. Biophys. J. 82, 1050–1058 (2002).
Xu, R. & Wilson, I. A. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J. Virol. 85, 5172–5182 (2011).
Huang, Q., Korte, T., Rachakonda, P. S., Knapp, E. W. & Herrmann, A. Energetics of the loop-to-helix transition leading to the coiled-coil structure of influenza virus hemagglutinin HA2 subunits. Proteins: Structure, Function, and Bioinformatics. 74, 291–303 (2009).
Basu, A. et al. New small molecule entry inhibitors targeting hemagglutinin-mediated influenza a virus fusion. J. Virol. 88, 1447–1460 (2014).
Antanasijevic, A. et al. Mutagenesis Studies of the H5 Influenza Hemagglutinin Stem Loop Region. J. Biol. Chem. 289, 22237–22245 (2014).
Luo, G., Colonno, R. & Krystal, M. Characterization of a Hemagglutinin-Specific Inhibitor of Influenza A Virus. Virology. 226, 66–76 (1996).
Staschke, K. A. et al. Inhibition of Influenza Virus Hemagglutinin-Mediated Membrane Fusion by a Compound Related to Podocarpic Acid. Virology. 248, 264–274 (1998).
Russell, R. J. et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. P. Natl. Acad. Sci. 105, 17736–17741 (2008).
Biasini, M. et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic. Acids. Res. gku340 (2014).
Guex, N., Peitsch, M. C. & Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 30, S162–S173 (2009).
Yang, H. et al. Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins. J. Virol. 88, 4828–4838 (2014).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R. & Thornton, J. M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 8, 477–486 (1996).
Gamblin, S. et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 303, 1838–1842 (2004).
Zhou, Y., Wu, C., Zhao, L. & Huang, N. Exploring the early stages of the pH‐induced conformational change of influenza hemagglutinin. Proteins: Structure, Function, and Bioinformatics. 82, 2412–2428 (2014).
Hess, B., Kutzner, C., Van Der Spoel, D. & Lindahl, E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory. Comput. 4, 435–447 (2008).
Zhan, D., Guan, S., Jin, H., Han, W. & Wang, S. Stereoselectivity of phosphotriesterase with paraoxon derivatives: a computational study. J. Biomol. Struct. Dyn. 34, 600–611 (2016).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Guan, S., Han, W., Zhang, H., Wang, S. & Shan, Y. Insight into the interactive residues between two domains of human somatic Angiotensin-converting enzyme and Angiotensin II by MM-PBSA calculation and steered molecular dynamics simulation. J. Biomol. Struct. Dyn. 34(1), 15–28 (2016).
Hess, B. & van der Vegt, N. F. Hydration thermodynamic properties of amino acid analogues: a systematic comparison of biomolecular force fields and water models. J. Phys. Chem. B. 110, 17616–17626 (2006).
Guan, S. et al. Binding modes of phosphotriesterase-like lactonase complexed with δ-nonanoic lactone and paraoxon using molecular dynamics simulations. J. Biomol. Struct. Dyn. 1–14 (2016).
Malde, A. K. et al. An Automated Force Field Topology Builder (ATB) and Repository: Version 1.0. J. Chem. Theory. Comput. 7, 4026–4037 (2011).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Berendsen, H. J., Postma, J. V., van Gunsteren, W. F., DiNola, A. & Haak, J. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Vorontsov, I. I. & Miyashita, O. Crystal molecular dynamics simulations to speed up MM/PB (GB) SA evaluation of binding free energies of di-mannose deoxy analogs with P51G-m4-Cyanovirin-N. J. Comput. Chem. 32, 1043–1053 (2011).
Jogalekar, A. S., Reiling, S. & Vaz, R. J. Identification of optimum computational protocols for modeling the aryl hydrocarbon receptor (AHR) and its interaction with ligands. Bioorg. Med. Chem. Lett. 20, 6616–6619 (2010).
Zhou, X., Xi, W., Luo, Y., Cao, S. & Wei, G. Interactions of a Water-Soluble Fullerene Derivative with Amyloid-β Protofibrils: Dynamics, Binding Mechanism, and the Resulting Salt-Bridge Disruption. J. Phys. Chem. B. 118, 6733–6741 (2014).
Case, D. A. et al. The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 (2005).
Fu, G., Liu, H. & Doerksen, R. J. Molecular modeling to provide insight into the substrate binding and catalytic mechanism of human biliverdin-IXα reductase. J. Phys. Chem. B. 116, 9580–9594 (2012).
Jin, H., Zhou, Z., Wang, D., Guan, S. & Han, W. Molecular Dynamics Simulations of Acylpeptide Hydrolase Bound to Chlorpyrifosmethyl Oxon and Dichlorvos. Int. J. Mol. Sci. 16, 6217–6234 (2015).
Réti-Nagy, K. et al. Endocytosis of fluorescent cyclodextrins by intestinal Caco-2 cells and its role in paclitaxel drug delivery. Int. J. Pharm. 496, 509–517 (2015).
Salmas, R. E., Yurtsever, M. & Durdagi, S. Investigation of inhibition mechanism of chemokine receptor CCR5 by micro-second molecular dynamics simulations. Sci. Rep. 5 (2015).
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & NA, B. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic acids research. 32(suppl 2), W665–W667 (2004).
Lin, X., Noel, J. K., Wang, Q., Ma, J. & Onuchic, J. N. Lowered pH Leads to Fusion Peptide Release and a Highly Dynamic Intermediate of Influenza Hemagglutinin. J. Phys. Chem. B. 120, 9654–9660 (2016).
Acknowledgements
The current work was supported by Key Projects of Science and Technology Bureau of Changchun City (Grant no. 14KG052), Industrial Technology Research and Development Projects of Jilin Province Development and Reform Commission (Grant no. 2014Y081), Science and Technology Enterprise Technology Innovation Fund by Jiangsu Province Science and Technology Department (Grant no. BC2015065), Project 2016008 Supported by the Graduate Innovation Fund of Jilin University, Science and Technology Research Project in 13th Five-Year Plan Period of Education Department of Jilin Province (Grant no. 2016406 and 2016408) and Scientific Frontier and Interdisciplinary Innovation Project Funding of Jilin University (Grant no. 2013ZY16).
Author information
Authors and Affiliations
Contributions
S.S.G. and Y.M.S. designed the experiments. S.S.G., T.A.W., X.P.T., X.Q.Z. and M.D.Q. carried out the experiments. S.S.G., Y.M.S., Z.Y.K., Y.J.Y., H.Z., S.W., H.L., and W.K. analyzed and discussed the results. S.S.G. wrote the manuscript. All authors have reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guan, S., Wang, T., Kuai, Z. et al. Exploration of binding and inhibition mechanism of a small molecule inhibitor of influenza virus H1N1 hemagglutinin by molecular dynamics simulation. Sci Rep 7, 3786 (2017). https://doi.org/10.1038/s41598-017-03719-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-017-03719-4
This article is cited by
-
Exploration of natural product database for the identification of potent inhibitor against IDH2 mutational variants for glioma therapy
Journal of Molecular Modeling (2023)
-
Computational biophysics approach towards the discovery of multi-kinase blockers for the management of MAPK pathway dysregulation
Molecular Diversity (2023)
-
Exploring safe and potent bioactives for the treatment of non-small cell lung cancer
3 Biotech (2021)
-
Mechanistic insights into the deleterious roles of Nasu-Hakola disease associated TREM2 variants
Scientific Reports (2020)
-
In silico structural elucidation of RNA-dependent RNA polymerase towards the identification of potential Crimean-Congo Hemorrhagic Fever Virus inhibitors
Scientific Reports (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.