Abstract
The worm-shaped, shell-less Caudofoveata is one of the least known groups of molluscs. As early-branching molluscs, the lack of high-quality genomes hinders our understanding of their evolution and ecology. Here, we report a high-quality chromosome-scale genome of Chaetoderma sp. combining PacBio, Illumina, and high-resolution chromosome conformation capture sequencing. The final assembly has a size of 2.45 Gb, with a scaffold N50 length of 141.46 Mb, and is anchored to 17 chromosomes. Gene annotations showed a high level of accuracy and completeness, with 23,675 predicted protein-coding genes and 94.44% of the metazoan conserved genes by BUSCO assessment. We further present 16S rRNA gene amplicon sequencing of the gut microbiota in Chaetoderma sp., which was dominated by the chemoautotrophic bacteria (phylum Gammaproteobacteria). This chromosome-level genome assembly presents the first genome for the Caudofoveata, which constitutes an important resource for studies ranging from molluscan evolution, symposium, to deep-sea adaptation.
Similar content being viewed by others
Background & Summary
The Aplacophora is a particular understudied molluscs that is evolutionarily and ecologically important in marine benthic fauna. As early-branching molluscs, Aplacophora is unusual as it is with worm-shaped, shell-less body plan, and covered by cuticle and calcareous sclerites. Two groups Caudofoveata and Solenogastres are often collectively referred to as Aplacophora1,2. The pedal groove does not exist in the ventral site of Caudofoveata, which distinguishes it from Solenogastres. Besides, Caudofoveata has gills at the tail of the body and is absence of a foot1. Caudofoveata has a worldwide distribution in benthic marine habitats and lives by burrowing in marine soft sediment feeding on organic contents or foraminiferans and small particles2,3. Due to the collection difficulty, Caudofoveata is one of the least known classes of Mollusca, with only 142 species (World Register of Marine Species, 2023). To data, a series of studies have researched their taxonomy4,5,6, phylogeny2,7,8,9,10,11, ecology3,12,13, and evolutions14,15,16,17.
Caudofoveata had been thought to be the earliest extant offshoots in Mollusca based on its unique body plan and shell-less morphological characters18,19. Phylogenomic analyses revealed that Mollusca included two clades, Aculifera (Caudofoveata, Solenogastres and polyplacophorans) and Conchifera (Gastropoda, Bivalvia, Cephalopoda, Scaphopoda and Monoplacophora)10,11. The fossil Kimberella quadrata was thought to be a stem-group mollusc and had certain traits similar to Aculifera, which indicated Caudofoveata was an early-branching Mollusca18,20,21. Overall, Caudofoveata is central to understand the origin and evolutionary history of molluscs, which is the second most diverse metazoan animal group.
Aplacophora is of particular interest from the benthic fauna of the deep sea especially regarding to its diversity and adaptation. With deep sea expeditions increasing in the Atlantic Ocean and the Northwest Pacific, more and more new Aplacophoran molluscs were described and studied3,22,23,24,25. Almost 86% of the Aplacophora were found at depths of more than 200 m1 and some species showed high abundances at deep-sea benthos26. Prochaetoderma yongei, a widespread deep sea Caudofoveata species in Atlantic, was thought to be successful due to its omnivorous and rapid development ability3,26. Helicoradomenia spp. which is a Solenogastres species has been found in the sulfide-based chemosynthetic hydrothermal vent with epi- and endocuticular bacterial symbionts27. By investigating the food sources and anatomy of 200 individuals within 60 candidate deep-sea Solenogastres species, researchers revealed a high degree of food specialization with modifications in the radula, foregut, and glands morphologies28. Considering their great ecological importance and diverse adaptation strategies to deep-sea environment, Aplacophora could be an ideal group to study deep-sea adaptation.
Despite the evolutionary and ecological importance, the studies of Caudofoveata are hampered by the lack of genomic resources. Here, we generated a high-quality chromosome-level genome of Chaetoderma sp. for the first time in the Caudofoveata based on PacBio long reads, Illumina short reads, and high-resolution chromosome conformation capture (Hi-C) sequencing reads. The final assembly of Chaetoderma sp. was 2.45 Gb, consisting of 17 chromosomes with scaffold N50 length of 141.46 Mb. We predicted 23,675 protein-coding genes from the genome of Chaetoderma sp. by integrating de-novo, homologous, and transcriptome annotation methods as well as manual correction. Through the analysis of intestinal microbial composition of Chaetoderma sp., we discovered that SUP05, a group of chemoautotrophic bacteria was the dominant bacterial community in the gut, indicating a potential symbiotic relationship between them. The resulting genome assembly, annotation, and report of symbiotic bacteria by 16S rRNA gene amplicon sequencing will provide a valuable resource for further studies of the Caudofoveata and for molluscan evolution and deep-sea ecology in general.
Methods
Sample collection and sequencing
The Chaetoderma sp. specimens were collected from Site F methane seep29 (also known as Formosa Ridge) by the TV grab in the South China Sea during the voyage of the scientific research ship KEXUE from 2020 to 2022. When the samples were collected by the TV grab onto the ship, they were flash-frozen in liquid nitrogen immediately and stored in −80 °C refrigerator. The same frozen specimen of Chaetoderma sp. was used to perform the Illumina, PacBio and Hi-C sequencing. The total genomic DNA was extracted from the body wall by SDS method and followed by chloroform purification, examination of the quantity and quality through Qubit and Agilent bioanalyzer instrument. The qualified genomic DNA was used to construct libraries.
Firstly, in order to estimate genome complexity, we used physical breaking method to break up the genome DNA to 350 bp fragment, and then built the small fragment sequencing library which was applied to an Novaseq 6000 platform (Illumina, Inc., San Diego, CA, USA). A total of 57.80 Gb 150 bp paired-end sequencing reads were obtained (Table 1). Secondly, the PacBio library was constructed by following the standard protocol of manufacturer (Pacific Biosciences, Menlo Park, CA, USA), including using g-TUBE to break up DNA, the repair of DNA, the connection of dumbbell connector, the digestion of exonuclease and the filtration of target DNA fragment by BluePippin. A total of three SMART cells and 62.4 Gb clean long HiFi reads with 26.01X coverage were sequenced through circular consensus sequencing (CCS) model (Table 1). Thirdly, we applied high-throughput chromatin conformation capture (Hi-C) method to generate a chromosome-level genome. As for Hi-C library, formaldehyde was used to fix cells, and DpnII restriction endonuclease was used to digest cells. Using the terminal repair mechanism, DNA was labelled and cycled. The Hi-C library was built by using streptavidin magnetic beads to selectively capture DNA fragments containing interaction relationships and was evaluated for quality through Qubit 2.0, Agilent 2100 systems and Q-PCR method. Illumina NovaSeq 6000 platform (Illumina, USA) was performed to execute Hi-C sequencing. We obtained 257.18 Gb clean reads in total (Table 1).
To better annotate the genome assembly, we performed transcriptome sequencing of Chaetoderma sp. using the frozen body. The total RNA was extracted using Trizol (Invitrogen). Qubit and agarose gel electrophoresis were further applied to examine the concentration and quality of RNA. VAHTS® Universal V8 RNA-seq Library Prep Kit for Illumina (Vazyme #NR605) was used to constructed RNA-seq library. The sequencing library was further sequenced on Novaseq 6000 platform in 150 bp paired-end mode (Illumina, Inc., San Diego, CA, USA). A total of 6.5 Gb raw reads were obtained.
Genome assembly and Hi-C scaffolding
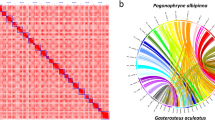
Genome size, repetitive sequence ratio, and heterozygosity were first estimated based on Illumina short-read data. We used jellyfish v2.3.030 and GenomeScope v1.031 to analyse the K-mer frequency (k = 21). Based on Illumina reads, the K-mer analysis showed that the genome size of Chaetoderma sp. was 2.2 Gb and the heterozygosity was 1.39%. HiFi-asm v0.1632 was applied to assemble the genome based on PacBio long-read data. Pilon v1.2333 was then used to polish the assembly with the Illumina short-read data. Purge_dups v1.2.534 was used to remove duplication. The assembly of Chaetoderma sp. genome is 2.45 Gb, consisting of 5,603 contigs with contig N50 of 1.06 Mb (Table 2). The BUSCO assessment value is 92.03% (metazoan_odb10) and the GC content of the genome assembly is 40.93%. As for Hi-C scaffolding, at first Juicer v1.635 was used to deal with the Hi-C sequencing data and obtain the input file for the next analysis. Then, 3D-DNA v20100836 as the core software was used to scaffold the contigs under default settings. Juicebox v1.11.0835 was used to visualize chromosome assembly results, choosing the best result from 3D-DNA output, and marking the correct boundary of the chromosome according to the interaction heatmap. Finally, we reran 3D-DNA using corrected assembly result and exported the final chromosome assembly genome. After Hi-C scaffolding, 94.83% genome reads were anchored to 17 chromosomes with scaffold N50 length of 141.46 Mb and BUSCO assessment value of 89.52% (Fig. 1, Tables 2, 4, and 6). The 17 chromosomes were exhibited clearly in the interaction heatmap (Fig. 2) and also had a conserved collinearity relationship with the chromosomes of Mizuhopecten yessoensis (Fig. 3). All the bioinformatic software mentioned in this section were used with default parameters.
Characterization of the assembled Chaetoderma sp. genome. From inner to outer layers: photograph of Chaetoderma sp.; distribution of GC content in the genome; repeat sequence density across the genome; LTR density across the genome; gene density across the genome. All the tracks are drawn in 1 Mb sliding windows and chromosome-level scaffolds at scale by circos v0.69-965.
Annotation of Repetitive Elements
De novo repeat library prediction and homology comparison were applied for repeats annotation. We employed RepeatModeler2 v2.0.137 with default parameters to construct the de novo repeat library. LTR_FINDER v1.0738 and LTR_retriever v2.9.039 were used to identify long terminal repeat (LTR) sequence in the genome by using default parameters. The de novo repeat library and LTR library were combined and removed redundancy to generate the final repeat library. RepeatMasker v4.140 (-frag 100000 -gc 33.37 -lcambig -xsmall -gff) was applied to identify repeats with RepBase and de novo species-specific library in the genome of Chaetoderma sp. The proportion of Transposon elements (TEs) in Chaetoderma sp. genome is 55.81%. Among them, retroelement accounts for 38.43%, DNA transposon accounts for 17.38%. The most abundant transposon type is the LTR (Table 5).
Gene Prediction
The Caudofoveata gene prediction is challenging because of the high ratios of TEs and long introns, as gene prediction programmes may split a single gene into truncated partial-gene models. We employed three different approaches to predict protein-coding genes, homolog-based, transcriptome-based annotation, and ab initio gene prediction. Homolog-based annotation was performed by TBLASTN v2.13.041 (-evalue 1e-10) based on homology sequences from Acanthopleura granulate, Crassostrea gigas. Genewise v2.4.142 (-nosplice_gtag -pretty -pseudo -gff -cdna -trans) was used to predict genes based on homologous proteins. Second, we fully utilized and integrated transcriptome evidence in the gene prediction process, since this evidence can be helpful in the case of high ratios of TEs and long introns. Trinity v2.13.243 was used for transcriptomic level de novo assembly with default parameters. Hisat2 v2.2.144 (--skip 8 --qc-filter) was used to align transcriptome data to the genome, StringTie v2.2.145 was used to predict the structure of all transcribed reads. Subsequently, Program to Assemble Spliced Alignment (PASA) v2.5.246 was employed to integrate genome and transcriptome results. Third, ab initio gene prediction was carried out on the repeat-masked genome assembly by Braker2 v2.1.647 and Augustus v3.5.048 with default parameters. Finally, EvidenceMolder v1.1.149 was employed to integrate gene models from different prediction tools. We further used tRNAscan-SE v1.3.150 and barrnap v0.9 (http://lup.lub.lu.se/student-papers/record/8914064) to identify tRNA and rRNA by using default parameters. Finally, we predicted 23,675 protein-coding genes from the chromosome-level genome of Chaetoderma sp. by integrating de novo, homologous, and transcriptome annotation methods as well as modification of several genes’ structure such as Hox by comparing with homolog species one by one manually (Table 3). We used the BUSCO v551 to evaluate the quality of annotation results. The BUSCO completeness score is 94.44%, and the single copy score is 91.82% (Table 4). 23,503 (99.27%) of the protein-coding genes we predicted were annotated through blasting against SwissProt52 and interproscan53 against pfam54 database (Table 3).
16S rRNA sequencing and analysis
The total genome DNA of the intestinal contents of Chaetoderma sp. was extracted through SDS method. After monitoring the DNA concentration and purity based on 1% agarose gels, DNA was diluted to 1 ng/µL using sterile water. Specific primer (V4: 515F-806R) and barcodes were applied to amplify 16S rRNA genes. The library was sequenced on the Illumina NovaSeq platform to obtain 250 bp paired-end reads. We used FLASH v1.2.1155 to merge paired-end reads and used fastp v0.20.056 for data quality control with default parameters. QIIME2 v20200657 with default parameters was used to obtain ASVs (Amplicon Sequence Variants) and annotate species based on Silva Database. The result of 16S rRNA sequencing showed that SUP05 was the most abundant bacteria in the intestinal contents of Chaetoderma sp. and SUP05 is a group of Gammaproteobacteria with chemoautotrophic ability58 (Fig. 4).
Data Records
The raw Illumina, PacBio, and Hi-C sequencing data are deposited in the NCBI under the accession number SRP45722559. The assembled genome sequence is deposited into NCBI under accession number GCA_034401795.160. The genome annotation file is available from the Figshare repository61. The SRA database of transcriptome data is SRR2694995462. The SRA database of raw Illumina 16S rRNA sequencing is under the accession number SRP45864763.
Technical Validation
Evaluating genome assembly and annotation completeness
The final assembly of Chaetoderma sp.’s genome is 2.45 Gb, consisting of 17 chromosomes with contig N50 of 1.06 Mb and scaffold N50 of 141.46 Mb (Fig. 1, Table 2). The genome size is similar with the result that was estimated by jellyfish. In order to evaluate the genome assembly and annotation, we adopted two methods including Illumina reads remapping using Bowtie2 v2.4.564 and BUSCO v551 assessment using database metazoan_odb10. The alignment rate of Illumina reads was 95% (Table 2). 854 (89.52%) of 954 BUSCOs were included in the assembly of Chaetoderma sp. and 901 (94.44%) of 954 BUSCOs were included in the gene models of Chaetoderma sp. (Table 4). We also compared our results with other molluscs’ assembly and annotation (Table 7). Overall, these data indicate the genome assembly and annotation of Chaetoderma sp. is complete and high-quality.
Code availability
No custom script was used in this work. Software that was used to analyse data was listed in methods in detail and commands were used based on the manuals.
References
Todt, C. Aplacophoran Mollusks—still obscure and difficult?*. Amer. Malac. Bull. 31, 181–187 (2013).
Mikkelsen, N. T., Todt, C., Kocot, K. M., Halanych, K. M. & Willassen, E. Molecular phylogeny of Caudofoveata (Mollusca) challenges traditional views. Mol. Phylogenet. Evol. 132, 138–150 (2019).
Scheltema, A. H. & Ivanov, D. L. A natural history of the deep-sea aplacophoran Prochaetoderma yongei and its relationship to confamilials (Mollusca, Prochaetodermatidae). Deep Sea Res. Part II Oceanogr. Res. Pap. 56, 1856–1864 (2009).
Passos, F. D., Corrêa, P. V. F. & Todt, C. A new species of Falcidens (Mollusca, Aplacophora, Caudofoveata) from the southeastern Brazilian coast: external anatomy, distribution, and comparison with Falcidens caudatus (Heath, 1918) from the USA. Mar. Biodiv. 48, 1135–1146 (2018).
Saito, H. & v. Salvini-Plawen, L. Four new species of the aplacophoran class Caudofoveata (Mollusca) from the southern Sea of Japan. J. Nat. Hist. 48, 2965–2983 (2014).
Señarís, M. P., García-Álvarez, O. & Urgorri, V. Four new species of Chaetodermatidae (Mollusca, Caudofoveata) from bathyal bottoms of the NW Iberian Peninsula. Helgoland Mar. Res. 70, 1-23 (2016).
Kocot, K. M., Todt, C., Mikkelsen, N. T. & Halanych, K. M. Phylogenomics of Aplacophora (Mollusca, Aculifera) and a solenogaster without a foot. Proc. Biol. Sci. 286, 1902 (2019).
Osca, D., Irisarri, I., Todt, C., Grande, C. & Zardoya, R. The complete mitochondrial genome of Scutopus ventrolineatus (Mollusca: Chaetodermomorpha) supports the Aculifera hypothesis. BMC Evol. Biol. 14, 197 (2014).
Mikkelsen, N. T., Kocot, K. M. & Halanych, K. M. Mitogenomics reveals phylogenetic relationships of caudofoveate aplacophoran molluscs. Mol. Phylogenet. Evol. 127, 429–436 (2018).
Kocot, K. M. et al. Phylogenomics reveals deep molluscan relationships. Nature 477, 452–456 (2011).
Smith, S. A. et al. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364–367 (2011).
Corrêa, P. V. F., Miranda, M. S. & Passos, F. D. South America-Africa missing links revealed by the taxonomy of deep-sea molluscs: Examples from prochaetodermatid aplacophorans. Deep Sea Res. Part I Oceanogr. Res. Pap. 132, 16–28 (2018).
Señarís, M. P., García-Álvarez, O. & Urgorri, V. The habitus of Scutopus robustus Salvini-Plawen, 1970 (Caudofoveata, Limifossoridae), a rare mollusc from the NW Iberian Peninsula. Mar. Biodivers. 47, 377–378 (2017).
Vinther, J., Sperling, E. A., Briggs, D. E. & Peterson, K. J. A molecular palaeobiological hypothesis for the origin of aplacophoran molluscs and their derivation from chiton-like ancestors. Proc. Biol. Sci. 279, 1259–1268 (2012).
Scherholz, M., Redl, E., Wollesen, T., Todt, C. & Wanninger, A. Aplacophoran mollusks evolved from ancestors with polyplacophoran-like features. Curr. Biol. 23, 2130–2134 (2013).
McDougall, C. & Degnan, B. M. The evolution of mollusc shells. Wires Dev. Biol. 7, e313 (2018).
Telford, M. J. Mollusc Evolution: Seven shells on the sea shore. Curr. Biol. 23, R952–R954 (2013).
Wanninger, A. & Wollesen, T. The evolution of molluscs. Biol. Rev. Camb. Philos. Soc. 94, 102–115 (2019).
Salvini-Plawen, L. v. & Steiner, G. The Testaria concept (Polyplacophora+Conchifera) updated. J. Nat. Hist. 48, 2751–2772 (2014).
Gehling, J. G., Runnegar, B. N. & Droser, M. L. Scratch Traces of Large Ediacara Bilaterian Animals. J. Paleontol. 88, 284–298 (2015).
Vinther, J. The origins of molluscs. J. Paleontol. 58, 19–34 (2015).
Cobo, M. C. & Kocot, K. M. On the diversity of abyssal Dondersiidae (Mollusca: Aplacophora) with the description of a new genus, six new species, and a review of the family. Zootaxa 4933, 63–97 (2021).
Bergmeier, F. S. et al. Of basins, plains, and trenches: Systematics and distribution of Solenogastres (Mollusca, Aplacophora) in the Northwest Pacific. Prog. Oceanogr. 178 (2019).
Cobo, M. C. & Kocot, K. M. Micromenia amphiatlantica sp. nov.: First solenogaster (Mollusca, Aplacophora) with an amphi-Atlantic distribution and insight into abyssal solenogaster diversity. Deep Sea Res. Part I Oceanogr. Res. Pap. 157 (2020).
Bergmeier, F. S., Brandt, A., Schwabe, E. & Jörger, K. M. Abyssal Solenogastres (Mollusca, Aplacophora) from the Northwest Pacific: scratching the surface of deep-sea diversity using integrative taxonomy. Front. Mar. Sci. 4 (2017).
Scheltema, A. H. Aplacophoran molluscs: Deep-sea analogs to polychaetes. B. Mar. Sci. 60, 575–583 (1997).
Katz, S., Cavanaugh, C. M. & Bright, M. Symbiosis of epi- and endocuticular bacteria with Helicoradomenia spp. (Mollusca, Aplacophora, Solenogastres) from deep-sea hydrothermal vents. Mar. Ecol. Prog. Ser. 320, 89–99 (2006).
Bergmeier, F. S., Ostermair, L. & Jorger, K. M. Specialized predation by deep-sea Solenogastres revealed by sequencing of gut contents. Curr. Biol. 31, R836–R837 (2021).
Feng, D. et al. Cold seep systems in the South China Sea: An overview. J. Asian Earth Sci. 168, 3–16 (2018).
Marcais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Vurture, G. W. et al. GenomeScope: fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Walker, B. J. et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9 (2014).
Guan, D. et al. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36, 2896–2898 (2020).
Durand, N. C. et al. Juicer Provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA 117, 9451–9457 (2020).
Xu, Z. & Wang, H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 35, W265–W268 (2007).
Ou, S. & Jiang, N. LTR_retriever: A highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018).
Tarailo-Graovac, M. & Chen, N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics Chapter 4, 4.10.1–4.10.14 (2009).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Birney, E., Clamp, M. & Durbin, R. GeneWise and genomewise. Genome Res. 14, 988–995 (2004).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–U130 (2011).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–U121 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Bruna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. Nar. Genom. Bioinform. 3, lqaa108 (2021).
Stanke, M. & Morgenstern, B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 33, W465–W467 (2005).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008).
Lowe, T. M. & Eddy, S. R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964 (1997).
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Apweiler, R. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Finn, R. D. et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285 (2016).
Magoc, T. & Salzberg, S. L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890 (2018).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Walsh, D. A. et al. Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326, 578–582 (2009).
NCBI sequence read archive https://identifiers.org/ncbi/insdc.sra:SRP457225 (2023).
Z, L. Chaetoderma sp. isolate LZ-2023a, whole genome shotgun sequencing project. Genbank https://identifiers.org/ncbi/insdc.gca:GCA_034401795.1 (2023).
Z, L. The annotation file of the chromosome-level genome of Chaetoderma sp. Figshare. https://doi.org/10.6084/m9.figshare.24099477 (2023).
NCBI sequence read archive https://identifiers.org/ncbi/insdc.sra:SRR26949954 (2023).
NCBI sequence read archive https://identifiers.org/ncbi/insdc.sra:SRP458647 (2023).
Langdon, W. B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 8, 1 (2015).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Acknowledgements
This research was supported by the Marine S & T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2022QNLM030004), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB42000000), National Natural Science Foundation of China (42376092 & 41976088), and Strategic Priority Research Program of the Chinese Academy of Sciences (XDA22050303). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Chunyu Zhang for helping with sample dissection. We thank Oceanographic Data Center, IOCAS; Qingdao Marine Science and Technology Center; as well as China Science and Technology Cloud for support of data analysis. We thank the vessels of “KEXUE” for help in sample collection.
Author information
Authors and Affiliations
Contributions
Conceived, designed, and supervised experiments: L.Z. Sample collection: L.Z. & M.W. Sample taxonomy: J.Z. Data collection: Y.W. & L.Z., Data analyses: Y.W. & J.L., Computer resources: L.Z. Wrote the paper: Y.W. & L.Z. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Y., Wang, M., Li, J. et al. A chromosome-level genome assembly of a deep-sea symbiotic Aplacophora mollusc Chaetoderma sp.. Sci Data 11, 133 (2024). https://doi.org/10.1038/s41597-024-02940-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-02940-x