Abstract
Batillaridae is a common gastropod family that occurs abundantly in the shallow coastal zone of the intertidal mudflats of the northwest Pacific Ocean, Australasia, and North America. In this family, Batillaria attramentaria is known for its biological invasion and colonization in estuarine and intertidal zones. It can endure and adapt the harsh intertidal conditions such as frequent temperature alteration, salinity, and air exposure. Therefore, we sequenced and assembled this Korean batillariid genome to get insight into its intertidal adaptive features. Approximately 53 Gb of DNA sequences were generated, and 863 scaffolds were assembled into a draft genome of 0.715 Gb with 97.1% BUSCO completeness value. A total of 40,596 genes were predicted. We estimated that B. attramentaria and Conus consors diverged about 230 million years ago (MYA) based on the phylogenetic analysis of closely related gastropod species. This genome study sets the footstep for genomics studies among native and introduced Batillaria populations and the Batillaridae family members.
Similar content being viewed by others
Background & Summary
Batillariidae, also called batillariids or mud-creepers are widely distributed in the north-western Pacific region of Asia along the complex coastline formed in Japan, Korea, eastern China, and America1,2,3,4. Within the Batillaridae family, B. attramentaria (Sowerby, 1855) is characterized by its habitats being limited to narrow intertidal zones consisting of rocks or sandy mud along coastlines and limited dispersal capacity associated with direct larval development5,6. Due to such biological constraints, geographical movement distance is limited, and its population structure is also inferred to be influenced by geographical factors6,7,8. These characteristics hinder them from escaping from their originated habitats. However, in the early 20th century, B. attramentaria was introduced into the Bay of the Northeast Coast in the United States and Canada by commercial shipment of oyster (Crassostrea gigas) aquaculture from Japan9,10. In the new habitat, this invaded species not only flourished but also successfully competed with the native gastropod species such as Cerithidea californica11,12,13,14.
The mitochondrial lineage of B. attramentaria is primarily subdivided into two, and their geographical distribution matches the trajectories of two dominant regional seawater currents, Tsushima and Kuroshio, that flow separately north and south of the Japanese archipelago2. An analysis of the demographic history of B. attramentaria indicates that this species has sharply increased approximately since the last glacial maximum (LGM: 26,000–19,000 years ago), directly influenced by the sea level rise and range expansion of habitat in Asia following climate change1.
Benthic organisms living in the estuarine intertidal zone are subjected to the most dynamic environmental circumstances, with frequently altered salinity and temperature in their habitat due to tidal conditions. Thus, estuarine intertidal organisms are continuously exposed daily to the submerged saline seawater and cold temperature during high tide and to the dry, low salinity and high temperature during the low tide. Subsequently, continuous exposure to such highly variable environmental conditions has shaped intertidal communities’ behavioural and physiological adaptation and genetic variation15,16. Salt stress exposure study on B. attramentaria shows that variation in salinity affects their locomotion activity17, which seems to be a typical response observed in several intertidal gastropods18,19,20. Among several studies of molluscs, a survey on intertidal oyster Crassostrea gigas highlights the pathways and genes involved in responding to and adapting to typical tidal environmental conditions17. In comparison, a study on terrestrial giant African snails shows the expansion of mucus-related gene families to mitigate dry conditions on the land and the doubling of several genes, including haemocyanin (a copper-containing respiratory protein) that helps in transporting oxygen and phosphoenolpyruvate carboxykinase gene families during whole genome duplication21. Adaptation to such typical intertidal and terrestrial environmental conditions was achieved by regulating water balance, air-breathing, nitrogen excretion, neural–immune system interactions, and specific behaviours.
In this context, the genome sequence of B. attramentaria will be beneficial for a deepened understanding of its evolution and invasiveness. It could be a suitable model for studying the combined influence of climate change and palaeoceanographic change on marine gastropods and other coastal taxa in the Northeast Asian region. As well as this study will enrich our knowledge of the genetic features involved in the adaptation to typical intertidal environmental factors.
Here, we present a first draft of reference genome assembly for B. attramentaria constructed using long reads generated by the Pacific Biosciences (PacBio) DNA sequencing platform Sequel and short paired-end reads generated by Illumina. The genome was assembled into 863 scaffolds (N50 = 1.28 Mb), with a total size of 0.715 Gb, with 97.2% assembly completeness analysed by BUSCO. The genome completeness is on par with the mollusc genomes sequenced to date. Structural annotation of the genome yielded 40,596 genes. Of the total genes predicted, 15,755 genes were functionally annotated with InterProScan. Based on phylogenetic analysis of related gastropod species, B. attramentaria diverged from Conus consors during the Early Mesozoic era, i.e., about 230 MYA. We have detected genes responsible for adapting to intertidal environments22 (Supplementary Table 1) such as the Na+/H+ exchanger family, Na+/K+ ATPase (for ionic regulation), acyltransferase, proline dehydrogenase (for osmotic regulation), haemocyanin beta-sandwich, animal haem peroxidase, protein-tyrosine phosphatase (for improving terrestrial respiratory function), and galactosyltransferase, Ependymin, TNF(Tumour Necrosis Factor) family, C1q domain (for immune defense), as observed in terrestrial and marine gastropods in previous studies15,16,21,22.
Methods
Sample collection and purification of DNA
To construct a draft of the reference genome for the Korean batillariids, we collected samples from B. attramentaria (Sowerby, 1855) from Hajeon-ri, Cheollabuk-do, South Korea (on November 2018 at 35°32′N, 126°33′E). The samples were kept alive in seawater during the transportation to the laboratory. To obtain high quality and molecular weight of DNA, we dissected fresh tissues from the foot to muscle part of the alive samples and quickly froze them at −80 °C. We did not include the gut part to avoid the snail’s intestinal microbiome contaminant to the snail DNA. Genomic DNA was extracted using the Dneasy® Blood & Tissue kit (Qiagen, Hilden, Germany), and the integrity was checked using an agarose gel.
Short-read DNA sequencing and genome size estimation
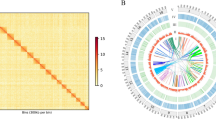
We constructed a library with an insert size of 350 bp using a Truseq Nano DNA Library kit (Illumina, SD, USA) following random fragmentation and adaptor ligation to DNA sequences. Paired-end (PE) sequencing with 101 bp was carried out using the Hiseq. 4000 sequencing system (Illumina, CA, USA), which generated a total of 731,221,132 PE reads (73.9 Gbp) (Supplementary Table 2). The JELLYFISH tool23 was used to estimate the genome size of B. attramentaria, which resulted in approximately 0.64 Gbp based on k-mer distribution value (K = 61). The main peak at k-mer depth 34 was used for genome size estimation (Fig. 1).
PacBio sequencing
The genomic DNA was sheared to generate ~20Kb fragments using the Covaris g-TUBE (Covaris) according to the manufacturer’s instructions. Small fragments were removed by the AMpureXP bead purification system (Beckman Coulter). A total of 5 μg DNA for each sample was used to prepare the library using SMRTbell® Express Template Prep Kit v2.0 (Pacific Biosciences, Menlo Park, CA, USA). Small fragments were removed from the library by BluePippin Size selection system for the large-insert library. Then the sequencing primer v4 was annealed to the SMRTbell template, followed by the binding of DNA polymerase to the complex (Sequel Binding kit 3.0). The excess primer and polymerase were removed from the complex using AMPure purification system before sequencing. Finally, the SMRT library was sequenced using the PacBio Sequel System with the Sequel Sequencing Kit v3.0 chemistry. A total of ~53.3 Gbp of subreads were obtained (Supplementary Table 3).
Genome assembly and polishing
Initially, cleaned PacBio long-read sequences were assembled using FALCON-Unzip assembler24, which generated a contiguous assembly of 844 Mbp (N50 = 1.08 Mbp). The larger size of the assembly than the estimated genome size suggested a high number of duplicate haplotypes25. The highly heterozygous genome assembly was curated by Purge Haplotigs26 to generate a de-duplicated haploid genome assembly. Further, the assembled genome was polished by Pilon 1.2.3 (with default parameters)27 by using aligned Illumina PE reads (57.5 Gb), resulting in a final assembly of 863 contigs with a total length of 715 Mb and an N50 length of 1.28 Mb (Table 1).
The assembled genome is much smaller than the closest sequenced genome of Conus consors (3.025 Gb)28. Due to high heterozygosity levels and repetitiveness, the assembly processes of molluscs are found to be complicated. Such instances were observed in oysters and other invertebrates29. The repeat content was estimated to be (314 Mb) 43.87% of the genome assembled (Table 2). Most invertebrate genomes, including molluscs, exhibit high heterozygosity and repetitiveness, complicating the assembly process. Genome completeness estimated by using BUSCO (Benchmarking Universal Single-Copy Orthologs) v3.0.2 detected a total of 927 (97.2%) of the 954 genes in the metazoan gene set30 (Table 3). Genome completeness is par with other mollusc genome assembly available till date (Fig. 2b)
(a) Divergence time tree among gastropods. Divergent times were estimated using RelTime methods with an ML phylogenomic tree, and the unit of time was scaled in millions of years. Bars around each node represent 95% confidence intervals. (b) Genome assembly completeness comparison estimated by BUSCO (metazoan_odb10). The genome assembly accession number is mentioned next to the plot. Assembly of A. purpuratus was analyzed from http://gigadb.org/dataset/100419. Star marks represent the chromosome-level assemblies.
Gene prediction and annotation
Before predicting genes, transposable elements (TEs) in the genome were identified using homology-based (RepeatMasker31, RepeatScout32, RepBase33, and RMBlast34) and by de novo using RepeatModeler35. Tandem Repeats Finder36 was used to predict consensus sequences and to gain classification information for each repeat. Annotation of repetitive elements resulted in 313,966,700 bp of repetitive DNA, amounting to 43.87% of the genome assembly (Table 3). The majority of the repetitive elements were unclassified (20.41%), followed by Simple repeats (7.38%), SINEs (5.55%), LINEs (4.83%), and DNA elements (3.81%). By using the SSRMMD tool37, we identified 1,518,868 simple sequence repeats (SSRs) distributed throughout the genome (Supplementary Table 4). A total of 3,304,085 SNPs has been detected in the B. attramentaria genome (Supplementary Table 5), after aligning sequence reads with the BWA tool38 and using bcftools39 to identify variants. Repetitive elements in the genome were masked before proceeding with the gene prediction. We used EvidenceModeller gene predicting tool for predicting protein-coding genes from the draft genome by combining evidence from ab initio gene predictions, transcripts, and protein homologues. We used Augustus40 for ab initio gene prediction. Additional supports for gene prediction came from two different data sets of transcripts generated by Trinity41 from our previous study by Ho et al.22 and homologous protein sequences of related species to B. attramentaria by PASA42 and Exonerate43. Finally, we used EVidenceModeller42 to merge and improve the ab initio predictions with the evidence of transcripts and protein sequences with weights of evidences. The predicted genes were annotated using InterProScan with Pfam44. A sensitive HMM scanning method on the known Pfam functional domains with an e-value of 0.05 was also used to classify the gene families. Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation was performed using the KEGG Automatic Annotation Server (https://www.genome.jp/kegg/kaas/)45 with the bi-directional best hit (BBH) method. Homology-based and ab initio-based gene prediction resulted in the identification of 40,596 protein-coding genes (i.e., a total of 29.8% of the genome) with an average gene length of 5,248 bp from the B. attramentaria genomes (Table 4). Functional annotation of all predicted protein-coding genes by InterpRoscan resulted in 15,756 (38.8%) genes by Pfam and 17,922 (44.1%) genes by Gene Ontology46. A total of 11,074 (27.3%) genes were annotated by KEGG database46.
Phylogenomics
We performed an extensive comparison of orthologous genes among 19 gastropod genomes (Batillaria attramentaria, Conus consors28, Lanistes nyassanus47, Marisa cornuarietis47, Pomacea canaliculata47, Aplysia californica, Elysia chlorotica48, Plakobranchus ocellatus49, Biomphalaria glabrata50, Bulinus truncates51, Achatina immaculata21, Lottia gigantea52, Chrysomallon squamiferum53, Haliotis rubra54, Crassostrea gigas55, Agropecten purpuratus56, and Octopus bimaculoides57) using OrthoFinder v3.058. With all species present, 3,532 orthogroups were formed, with 36 of those consisting of one-copy genes. With the fasttree tool provided in OrthoFinder, we constructed a tree of rooted species using 573 orthogroups, where at least 81.8% of species had single-copy genes in any orthogroup with Octopus bimaculoides as the outgroup. Divergence time was calculated using the species tree generated by using RelTime methods in MEGA-X59 with the Jones–Taylor–Thornton model (Fig. 2a). The timetree was computed using two calibration constraints with confidence interval (CI) of Haliotis rubra–Chrysomallon squamiferum (414–596.9 MYA) and of Elysia chlorotica–Aplysia californica (58.3–278.9 MYA) that were taken from the TimeTree database60 for the calibration of time trees. The divergence time between B. attramentaria and C. consors was approximately 230 MYA, i.e., during the Early Mesozoic era.
Comparative genomic analysis
A comparison of orthologous gene groups shared among related gastropods of C. consors, L. nyassanus, M. cornuarietis, and P. canaliculata analysis by OrthoVenn261 showed a core set of 5,679 gene groups and a unique set of 1,724 gene groups was specific to B. attramentaria (Fig. 3). Gene ontology (GO) enrichment analysis of the gene groups unique to B. attramentaria showed the top five over-representation of GO terms mostly related to protein poly-ADP-ribosylation, GTP binding, and innate immune response (Supplementary Table 6).
Data Records
All DNA and RNA raw reads have been deposited in the NCBI SRA. All short and long read DNA sequences are available under the NCBI SRA accession number SRP26999662, genome assembly with accession number GCA_018292915.163 and the whole genome shotgun sequencing project was deposited in GenBank accession JACVVK00000000064 under the BioProject no. PRJNA640962. Supplementary materials which include all supplementary tables, results of comparative genomics and phylogenomic analysed by OrthoFinder, SNPs and SSRs are deposited to Figshare repository46: https://doi.org/10.6084/m9.figshare.22309195.v4.
Technical Validation
Quality assessment of the DNA and purification
High-quality DNA with bands around and above 10 kb in the agarose gel was selected for sequencing. The quality of the genomic DNA was measured using Bioanalyzer 2100 (Agilent Technologies, CA, USA), and the quantity was measured by a NanoDrop-1000 microspectrophotometer.
Sequencing read quality validation
FastQC quality control (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) was used to assess the quality of raw high-throughput DNA sequencing datasets. Low-quality sequence PE-reads (<Q20) were filtered out by v.0.3265 before assembly46.
Gene prediction and annotation validation
Final gene model prediction of the B. attramentaria genome assembly were considered by Evidence Modeler and assessed with the BUSCO (metazoa_odb10). The final predicted gene set consisted of 40,596 genes with (Table 4) with BUSCO value of 81.6%.
Code availability
In this study, software tools used according to the description mentioned in the materials and method section. No custom code was used.
References
Ho, P. T., Kwan, Y. S., Kim, B. & Won, Y. J. Postglacial range shift and demographic expansion of the marine intertidal snail Batillaria attramentaria. Ecol. Evol. 5, 419–435 (2015).
Kojima, S., Hayashi, I., Kim, D., Iijima, A. & Furota, T. Phylogeography of an intertidal direct-developing gastropod Batillaria cumingi around the Japanese Islands. Mar. Ecol. Prog. Ser. 276, 161–172 (2004).
Ozawa, T., Köhler, F., Reid, D. G. & Glaubrecht, M. Tethyan relicts on continental coastlines of the northwestern Pacific Ocean and Australasia: molecular phylogeny and fossil record of batillariid gastropods (Caenogastropoda, Cerithioidea). Zool. Scr. 38, 503–525 (2009).
Prozorova1, L. A., Volvenko, I. E. & Noseworthy, R. Distribution and ecological morphs of northwestern Pacific gastropod Batillaria attramentaria (GB Sowerby II, 1855)(Cenogastropoda: Batillariidae). Marine Ecosystems under the Global Change in the Northwestern Pacific, 139 (2012).
Adachi, N. & Wada, K. Distribution in relation to life history in the direct–developing gastropod Batillaria cumingi (Batillariidae) on two shores of contrasting substrata. J. Molluscan Stud. 65, 275–288 (1999).
Adachi, N. Distribution of Batillaria cumingii (Gastropoda, Potamididae) in Tanabe Bay, middle Japan. Nankiseibutu 39, 33–38 (1997).
Whitlatch, R. & Obrebski, S. Feeding selectivity and coexistence in two deposit-feeding gastropods. Mar. Biol. 58, 219–225 (1980).
Yamada, S. B. Growth and longevity of the mud snail Batillaria attramentaria. Mar. Biol. 67, 187–192 (1982).
Bonnot, P. A recent introduction of exotic species of molluscs into California waters from Japan. Nautilus 49, 1–2 (1935).
Fisheries, U. S. B. O. & Galtsoff, P. S. Introduction of Japanese oysters into the United States. (1932).
Byers, J. E. The distribution of an introduced mollusc and its role in the long-term demise of a native confamilial species. Biol. Invasions 1, 339–352 (1999).
Byers, J. E. Competition between two estuarine snails: implications for invasions of exotic species. Ecology 81, 1225–1239 (2000).
Byers, J. E. Differential susceptibility to hypoxia aids estuarine invasion. Mar. Ecol. Prog. 203, 123–132 (2000).
Byers, J. E. Effects of body size and resource availability on dispersal in a native and a non-native estuarine snail. J. Exp. Mar. Biol. Ecol. 248, 133–150 (2000).
Zhang, G. et al. Molecular basis for adaptation of oysters to stressful marine intertidal environments. Annu. Rev. Anim. Biosci. 4, 357–381 (2016).
Zhang, Y. et al. Proteomic basis of stress responses in the gills of the pacific oyster Crassostrea gigas. J. Proteome Res. 14, 304–317 (2015).
Ho, P. T., Nguyen, H. Q., Kern, E. M. & Won, Y. J. Locomotor responses to salt stress in native and invasive mud‐tidal gastropod populations (Batillaria). Ecol. Evol. 11, 458–470 (2021).
Elfwing, T. & Tedengren, M. Effects of copper and reduced salinity on grazing activity and macroalgae production: a short-term study on a mollusc grazer, Trochus maculatus, and two species of macroalgae in the inner Gulf of Thailand. Mar. Biol. 140, 913–919 (2002).
Hughes, J., Chapman, H. & Kitching, R. Effects of sublethal concentrations of copper and freshwater on behaviour in an estuarine gastropod Polinices sordidus. Mar. Pollut. Bull. 18, 127–131 (1987).
Kitching, R., Chapman, H. & Hughes, J. Levels of activity as indicators of sublethal impacts of copper contamination and salinity reduction in the intertidal gastropod, Polinices incei Philippi. Mar. Environ. Res. 23, 79–87 (1987).
Liu, C. et al. Giant African snail genomes provide insights into molluscan whole‐genome duplication and aquatic–terrestrial transition. Mol. Ecol. Resour. 21, 478–494 (2021).
Ho, P.-T. et al. Impacts of salt stress on locomotor and transcriptomic responses in the intertidal gastropod Batillaria attramentaria. Biol. Bull. 236, 224–241 (2019).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Chin, C.-S. et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016).
Dias, G. B. et al. Haplotype-resolved genome assembly enables gene discovery in the red palm weevil Rhynchophorus ferrugineus. Sci. Rep. 11, 1–14 (2021).
Roach, M. J., Schmidt, S. A. & Borneman, A. R. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19, 1–10 (2018).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS one 9, e112963 (2014).
Andreson, R. et al. Gene content of the fish-hunting cone snail Conus consors. BioRxiv, 590695 (2019).
Powell, D. et al. The genome of the oyster Saccostrea offers insight into the environmental resilience of bivalves. DNA Res. 25, 655–665 (2018).
Seppey, M., Manni, M. & Zdobnov, E. M. in Gene prediction 227–245 (Springer, 2019).
Chen, N. Using Repeat Masker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics 5, 4.10. 11–14.10. 14 (2004).
Price, A. L., Jones, N. C. & Pevzner, P. A. De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358 (2005).
Jurka, J. et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110, 462–467 (2005).
Bedell, J. A., Korf, I. & Gish, W. MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics 16, 1040–1041 (2000).
Abrusán, G., Grundmann, N., DeMester, L. & Makalowski, W. TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25, 1329–1330 (2009).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Gou, X. et al. SSRMMD: A rapid and accurate algorithm for mining SSR feature loci and candidate polymorphic SSRs based on assembled sequences. Front. in Genetics 11, 706 (2020).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Stanke, M., Diekhans, M., Baertsch, R. & Haussler, D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644 (2008).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Haas, B. J. et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 9, 1–22 (2008).
Slater, G. S. C. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 1–11 (2005).
Punta, M. et al. The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 (2012).
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C. & Kanehisa, M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185 (2007).
Patra, A. K. et al. Genome assembly of the Korean intertidal mud-creeper Batillaria attramentaria. figshare https://doi.org/10.6084/m9.figshare.22309195.v4 (2023).
Sun, J. et al. Signatures of divergence, invasiveness, and terrestrialization revealed by four apple snail genomes. Mol. Biol. Evol. 36, 1507–1520 (2019).
Cai, H. et al. A draft genome assembly of the solar-powered sea slug Elysia chlorotica. Sci. Data 6, 1–13 (2019).
Maeda, T. et al. Chloroplast acquisition without the gene transfer in kleptoplastic sea slugs, Plakobranchus ocellatus. Elife 10, e60176 (2021).
Adema, C. M. et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 8, 15451 (2017).
Young, N. D. et al. Nuclear genome of Bulinus truncatus, an intermediate host of the carcinogenic human blood fluke Schistosoma haematobium. Nat. Commun. 13, 977 (2022).
Simakov, O. et al. Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526–531 (2013).
Sun, J. et al. The Scaly-foot Snail genome and implications for the origins of biomineralised armour. Nat. Commun. 11, 1–12 (2020).
Gan, H. M. et al. Best foot forward: Nanopore long reads, hybrid meta-Assembly, and haplotig purging optimizes the first genome assembly for the Southern Hemisphere blacklip abalone (Haliotis rubra). Front. in Genetics 10, 889 (2019).
Zhang, G. et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 (2012).
Li, C. et al. Draft genome of the Peruvian scallop Argopecten purpuratus. GigaScience 7, giy031 (2018).
Albertin, C. B. et al. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220–224 (2015).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14 (2019).
Mello, B. Estimating timetrees with MEGA and the TimeTree resource. Mol. Biol. Evol. 35, 2334–2342 (2018).
Hedges, S. B., Dudley, J. & Kumar, S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22, 2971–2972 (2006).
Xu, L. et al. OrthoVenn2: a web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 47, W52–W58 (2019).
Patra, A. K. et al. NCBI Sequence Read Archive. NCBI Sequence Read Archive https://identifiers.org/ncbi/insdc.sra:SRP269996 (2020).
Patra, A. K. et al. Genome assembly of Batillaria attramentaria EWHU_Batt_1.0. NCBI Genome https://identifiers.org/ncbi/insdc.gca:GCA_018292915.1 (2020).
Patra, A. K. et al. Batillaria attramentaria isolate Wonlab-2016, whole genome shotgun sequencing project. GenBank https://identifiers.org/ncbi/insdc:JACVVK000000000 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Acknowledgements
This study was supported by the (NRF) of Korea and the Korean government, Ministry of Science, Information and Communication Technology, and Future Planning of Korea (MSIP) (NFR-2015R1A4A1041997) to Y-J.W. and Y.K., and by Basic Science Research Program through the National Research Foundation of Korea(NRF) funded by the Ministry of Education (2019R1I1A2A02057134) to Y-J.W., and by a grant (No. PE9988B and PEA0083) of Korea Institute of Ocean Science & Technology, KIOST, to Y-J.W. This work was also supported by the Ewha Womans University Research Grant of 2023 to Y-J.W.
Author information
Authors and Affiliations
Contributions
Y.J.W. and Y.K. conceptualized the project. P.-T.H. and Y.-J.W. collected the sample. P.-T.H. purified DNA and RNA. S.J.L. sequenced, assembled the genome, and predicted gene model. A.K.P., P.-T.H. and S.J. analysed the data. A.K.P. and Y-J.W. wrote the manuscript. All authors edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Patra, A.K., Ho, PT., Jun, S. et al. Genome assembly of the Korean intertidal mud-creeper Batillaria attramentaria. Sci Data 10, 498 (2023). https://doi.org/10.1038/s41597-023-02403-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-023-02403-9