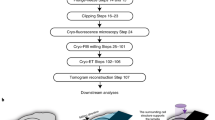
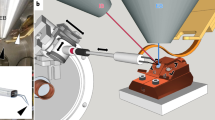
Abstract
Single-particle cryo-electron microscopy (cryoEM) provides an attractive avenue for advancing our atomic resolution understanding of materials, molecules and living systems. However, the vast majority of published cryoEM methodologies focus on the characterization of aerobically purified samples. Air-sensitive enzymes and microorganisms represent important yet understudied systems in structural biology. We have recently demonstrated the success of an anaerobic single-particle cryoEM workflow applied to the air-sensitive nitrogenase enzymes. In this protocol, we detail the use of Schlenk lines and anaerobic chambers to prepare samples, including a protein tag for monitoring sample exposure to oxygen in air. We describe how to use a plunge freezing apparatus inside of a soft-sided vinyl chamber of the type we routinely use for anaerobic biochemistry and crystallography of oxygen-sensitive proteins. Manual control of the airlock allows for introduction of liquid cryogens into the tent. A custom vacuum port provides slow, continuous evacuation of the tent atmosphere to avoid accumulation of flammable vapors within the enclosed chamber. These methods allowed us to obtain high-resolution structures of both nitrogenase proteins using single-particle cryoEM. The procedures involved can be generally subdivided into a 4 d anaerobic sample generation procedure, and a 1 d anaerobic cryoEM sample preparation step, followed by conventional cryoEM imaging and processing steps. As nitrogen is a substrate for nitrogenase, the Schlenk lines and anaerobic chambers described in this procedure are operated under an argon atmosphere; however, the system and these procedures are compatible with other controlled gas environments.
Key points
-
Structural analysis of air-sensitive enzymes requires that strictly anaerobic conditions be maintained during sample preparation and analysis. In this protocol, a customized vacuum manifold is used in the purification of nitrogenases from Azotobacter vinelandii and a custom anaerobic chamber is used for protein preparation for cryo-electron microscopy.
-
An internal oxygen probe made by genetically tagging the nitrogenase Fe protein with the far-red fluorescent protein mPlum can be expressed to report on oxygen exposure.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The single-particle cryoEM maps and models have been deposited into the PDB and Electron Microscopy Data Bank (EMDB) for release upon publication. Datasets been deposited with the following PDB and EMDB codes: 8TC3, EMD-41151 (Fe protein, mPlum tagged); 8DBY, EMD-27317 (MoFe protein on ultrathin carbon); 8DFC, EMD-27404 (ADP–AlF4− stabilized MoFe protein:Fe protein 1:1 complex); 8DFD, EMD-27405 (ADP–AlF4− stabilized MoFe protein:Fe protein 2:1 complex). All other data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Thamdrup, B. New pathways and processes in the global nitrogen cycle. Ann. Rev. Ecol. Evol. Syst. 43, 407–428 (2012).
Thorneley, R. & Lowe, D. Molybdenum enzymes. Met. Ions Biol. 7, 221–284 (1985).
Holland, P. L. Introduction: reactivity of nitrogen from the ground to the atmosphere. Chem. Rev. 120, 4919–4920 (2020).
Gallon, J. R. The oxygen sensitivity of nitrogenase: a problem for biochemists and micro-organisms. Trends Biochem. Sci. 6, 19–23 (1981).
Lowe, D. & Thorneley, R. N. The mechanism of Klebsiella pneumoniae nitrogenase action. The determination of rate constants required for the simulation of the kinetics of N2 reduction and H2 evolution. Biochem. J. 224, 895–901 (1984).
Buscagan, T. M., Kaiser, J. T. & Rees, D. C. Selenocyanate derived Se-incorporation into the nitrogenase Fe protein cluster. eLife 11, e79311 (2022).
Henthorn, J. T. et al. Localized electronic structure of nitrogenase FeMoco revealed by Selenium K-edge high resolution X-ray absorption spectroscopy. J. Am. Chem. Soc. 141, 13676–13688 (2019).
Spatzal, T., Perez, K. A., Howard, J. B. & Rees, D. C. Catalysis-dependent selenium incorporation and migration in the nitrogenase active site iron-molybdenum cofactor. eLife 4, e11620 (2015).
Sippel, D. et al. A bound reaction intermediate sheds light on the mechanism of nitrogenase. Science 359, 1484–1489 (2018).
Buscagan, T. M., Perez, K. A., Maggiolo, A. O., Rees, D. C. & Spatzal, T. Structural characterization of two CO molecules bound to the nitrogenase active site. Angew. Chem. Int. Ed. 60, 5704–5707 (2021).
Spatzal, T., Perez, K. A., Einsle, O., Howard, J. B. & Rees, D. C. Ligand binding to the FeMo-cofactor: structures of CO-bound and reactivated nitrogenase. Science 345, 1620–1623 (2014).
Einsle, O. & Rees, D. C. Structural enzymology of nitrogenase enzymes. Chem. Rev. 120, 4969–5004 (2020).
Nakane, T. et al. Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 (2020).
Yip, K. M., Fischer, N., Paknia, E., Chari, A. & Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161 (2020).
Warmack, R. A. et al. Structural consequences of turnover-induced homocitrate loss in nitrogenase. Nat. Commun. 14, 1091 (2023).
Rutledge, H. L., Cook, B. D., Nguyen, H. P. M., Herzik, M. A. & Tezcan, F. A. Structures of the nitrogenase complex prepared under catalytic turnover conditions. Science 377, 865–869 (2022).
Warmack, R. A. & Rees, D. C. Nitrogenase beyond the resting state: a structural perspective. Molecules 28, 7952 (2023).
Shriver, D. F. & Drezdzon, M. A. The Manipulation of Air-Sensitive Compounds 2nd edn (Wiley, 1986).
Lee, C. C., Ribbe, M. W. & Hu, Y. Purification of nitrogenase proteins. Methods Mol. Biol. 1876, 111–124 (2019).
Wiig, J. A., Lee, C. C., Fay, A. W., Hu, Y. & Ribbe, M. W. Purification of nitrogenase proteins. Methods Mol. Biol. 766, 93–103 (2011).
Jiménez-Vicente, E. et al. Application of affinity purification methods for analysis of the nitrogenase system from Azotobacter vinelandii. Methods Enzymol. 613, 231–255 (2018).
Spatzal, T. et al. Evidence for interstitial carbon in nitrogenase FeMo cofactor. Science 334, 940 (2011).
Kim, J., Woo, D. & Rees, D. C. X-ray crystal structure of the nitrogenase molybdenum–iron protein from Clostridium pasteurianum at 3.0-A resolution. Biochemistry 32, 7104–7115 (1993).
Mayer, S. M., Lawson, D. M., Gormal, C. A., Roe, S. M. & Smith, B. E. New insights into structure–function relationships in nitrogenase: a 1.6 A resolution X-ray crystallographic study of Klebsiella pneumoniae MoFe-protein. J. Mol. Biol. 292, 871–891 (1999).
Dos Santos, P. C. Molecular biology and genetic engineering in nitrogen fixation. Methods Mol. Biol. 766, 81–92 (2011).
Dos Santos, P. C. Genomic manipulations of the diazotroph Azotobacter vinelandii. Methods Mol. Biol. 1876, 91–109 (2019).
Echavarri-Erasun, C., Arragain, S. & Rubio, L. M. Purification of O2-sensitive metalloproteins. Methods Mol. Biol. 1122, 5–18 (2014).
Uchendu, S. N., Rafalowski, A., Cohn, E. F., Davoren, L. W. & Taylor, E. A. Anaerobic protein purification and kinetic analysis via oxygen electrode for studying DesB dioxygenase activity and inhibition. J. Vis. Exp. https://doi.org/10.3791/58307 (2018).
Uzarski, J. S., DiVito, M. D., Wertheim, J. A. & Miller, W. M. Essential design considerations for the resazurin reduction assay to noninvasively quantify cell expansion within perfused extracellular matrix scaffolds. Biomaterials 129, 163–175 (2017).
Shaner, N. C. et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 (2004).
Moore, M. M., Oteng-Pabi, S. K., Pandelieva, A. T., Mayo, S. L. & Chica, R. A. Recovery of red fluorescent protein chromophore maturation deficiency through rational design. PLoS ONE 7, e52463 (2012).
Linkerhägner, K. & Oelze, J. Cellular ATP levels and nitrogenase switchoff upon oxygen stress in chemostat cultures of Azotobacter vinelandii. J. Bacteriol. 177, 5289–5293 (1995).
Wenke, B. B., Arias, R. J. & Spatzal, T. Crystallization of nitrogenase proteins. Methods Mol. Biol. 1876, 155–165 (2019).
Wenke, B. B. The Many Roles of the Nitrogenase Iron Protein PhD thesis, California Institute of Technology (2019).
Chen, J., Noble, A. J., Kang, J. Y. & Darst, S. A. Eliminating effects of particle adsorption to the air/water interface in single-particle cryo-electron microscopy: bacterial RNA polymerase and CHAPSO. J. Struct. Biol. X. https://doi.org/10.1016/j.yjsbx.2019.100005 (2019).
Noble, A. J. et al. Routine single particle CryoEM sample and grid characterization by tomography. eLife https://doi.org/10.7554/eLife.34257 (2018).
Noble, A. J. et al. Reducing effects of particle adsorption to the air-water interface in cryo-EM. Nat. Methods 15, 793–795 (2018).
Passmore, L. A. & Russo, C. J. Specimen preparation for high-resolution cryo-EM. Methods Enzymol. 579, 51–86 (2016).
Wagner, A. O. et al. Medium preparation for the cultivation of microorganisms under strictly anaerobic/anoxic conditions. J. Vis. Exp. https://doi.org/10.3791/60155 (2019).
Lambertz, C. et al. O2 reactions at the six-iron active site (H-cluster) in [FeFe]-hydrogenase. J. Biol. Chem. 286, 40614–40623 (2011).
Gillman, C., Nicolas, W. J., Martynowycz, M. W. & Gonen, T. Design and implementation of suspended drop crystallization. Preprint at bioRxiv https://doi.org/10.1101/2023.03.28.534639 (2023).
Cherrier, M. V. et al. Oxygen-sensitive metalloprotein structure determination by cryo-electron microscopy. Biomolecules https://doi.org/10.3390/biom12030441 (2022).
Tivol, W. F., Briegel, A. & Jensen, G. J. An improved cryogen for plunge freezing. Microsc. Microanal. 14, 375–379 (2008).
Schmidt, F. V. et al. Structural insights into the iron nitrogenase complex. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-023-01124-2 (2023).
Fajardo, A. S. et al. Structural insights into the mechanism of the radical SAM carbide synthase NifB, a key nitrogenase cofactor maturating enzyme. J. Am. Chem. Soc. 142, 11006–11012 (2020).
Kang, W. et al. X-Ray crystallographic analysis of NifB with a full complement of clusters: structural insights into the radical SAM-dependent carbide insertion during nitrogenase cofactor assembly. Angew. Chem. Int. Ed. 60, 2364–2370 (2021).
Ferreira, C. M. H., Pinto, I. S. S., Soares, E. V. & Soares, H. M. V. M. (Un)suitability of the use of pH buffers in biological, biochemical and environmental studies and their interaction with metal ions—a review. RSC Adv. 5, 30989–31003 (2015).
Scheller, K. H. et al. Metal ion/buffer interactions. Eur. J. Biochem. 107, 455–466 (1980).
Zawisza, I., Rózga, M., Poznański, J. & Bal, W. Cu(II) complex formation by ACES buffer. J. Inorg. Biochem. 129, 58–61 (2013).
Wolle, D., Kim, C., Dean, D. & Howard, J. B. Ionic interactions in the nitrogenase complex. Properties of Fe-protein-containing substitutions for Arg-100. J. Biol. Chem. 267, 3667–3673 (1992).
Robinson, A. C., Burgess, B. K. & Dean, D. R. Activity, reconstitution, and accumulation of nitrogenase components in Azotobacter vinelandii mutant strains containing defined deletions within the nitrogenase structural gene cluster. J. Bacteriol. 166, 180–186 (1986).
Schlenk, W. & Thal, A. Über metallketyle, eine grosse klasse von verbindungen mit dreiwertigem kohlenstoff II. Ber. Dtsch. Chem. Ges. 46, 2840–2854 (1913).
Dilworth, M. J. Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim. Biophys. Acta 127, 285–294 (1966).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Thompson, R. F., Iadanza, M. G., Hesketh, E. L., Rawson, S. & Ranson, N. A. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat. Protoc. 14, 100–118 (2019).
Keable, S. M. et al. Structural characterization of the P(1+) intermediate state of the P-cluster of nitrogenase. J. Biol. Chem. 293, 9629–9635 (2018).
Danyal, K. et al. Negative cooperativity in the nitrogenase Fe protein electron delivery cycle. Proc. Natl Acad. Sci. USA 113, E5783–E5791 (2016).
Vénien-Bryan, C. & Fernandes, C. A. H. Overview of membrane protein sample preparation for single-particle cryo-electron microscopy analysis. Int. J. Mol. Sci. https://doi.org/10.3390/ijms241914785 (2023).
Acknowledgements
This work was funded by support from the Howard Hughes Medical Institute (D.C.R.), National Institutes of Health grants GM045162 (D.C.R.) and GM143836-01 (R.A.W.). The foundational contributions of J. Howard to establishing the anaerobic methodology within the laboratory are gratefully acknowledged. We thank J. Kaiser, S. Chen, A. Maggiolo and N. Siladke for their invaluable discussions. Azotobacter vinelandii DJ54 strain was a kind gift from D. Dean. The generous support of the Beckman Institute for the Caltech CryoEM Resource Center was essential for the performance of this research.
Author information
Authors and Affiliations
Contributions
T.S. and B.B.W. constructed and installed equipment. T.S., B.B.W. and R.A.W. established protocols. All authors contributed to the writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Warmack, R. A. et al. Nat. Commun. 14, 1091 (2023): https://doi.org/10.1038/s41467-023-36636-4
Key data used in this protocol
Warmack, R. A. et al. Nat. Commun. 14, 1091 (2023): https://doi.org/10.1038/s41467-023-36636-4
Supplementary information
Supplementary Information
Supplementary Figs. 1–4.
Source data
Source Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Warmack, R.A., Wenke, B.B., Spatzal, T. et al. Anaerobic cryoEM protocols for air-sensitive nitrogenase proteins. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-00973-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-00973-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.