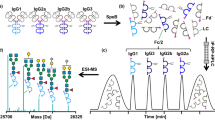
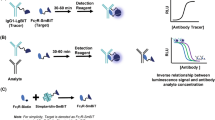
Abstract
Immunoglobulin G (IgG) fragment crystallizable (Fc) glycosylation modulates effector functions such as antibody-dependent cellular cytotoxicity and complement-dependent cytotoxicity. Consequently, assessing IgG Fc glycosylation is important for understanding the role of antibodies in infectious, alloimmune and autoimmune diseases. GlYcoLISA determines the Fc glycosylation of antigen-specific IgG by an immunosorbent assay with a liquid chromatography–mass spectrometry (LC–MS) readout. Detection of antigen-specific IgG glycosylation in a subclass- and site-specific manner is realized by LC–MS-based glycopeptide analysis after proteolytic cleavage. GlYcoLISA addresses challenges related to the low abundance of specific IgG and the high background of total IgG by using well-established immunosorbent assays for purifying antibodies of the desired specificity using immobilized antigen. Alternative methods with sufficient glycan resolution lack these important specificities. GlYcoLISA is performed in a 96-well plate format, and the analysis of 160 samples takes ~5 d, with 1 d for sample preparation, 2 d of LC–MS measurement and 2 d for partially automated data processing. GlYcoLISA requires expertise in LC–MS operation and data processing.
Key points
-
This protocol describes a method for profiling fragment crystallizable glycosylation of antigen-specific antibodies isolated from clinical samples.
-
By characterizing IgG specific for an antigen of interest and at a high molecular resolution, the technique allows more confident and functionally relevant characterization of glycosylation alterations in disease than could be achieved with less-specific methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
All data reported here (Figs. 3 and 5–7) have been previously reported elsewhere or are available in the source data: Fig. 3 and ref. 60 are available upon request from t.pongracz@lumc.nl; Fig. 5, source data; Fig. 6, ref. 20 and Supplementary Table 1; and Fig. 7, ref. 44 and Supplementary Table 1. Source data are provided with this paper.
References
Arnold, J. N., Wormald, M. R., Sim, R. B., Rudd, P. M. & Dwek, R. A. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu. Rev. Immunol. 25, 21–50 (2007).
de Haan, N., Falck, D. & Wuhrer, M. Monitoring of immunoglobulin N- and O-glycosylation in health and disease. Glycobiology 30, 226–240 (2020).
Ferrara, C. et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc. Natl Acad. Sci. USA 108, 12669–12674 (2011).
van Osch, T. L. J. et al. Fc galactosylation promotes hexamerization of human IgG1, leading to enhanced classical complement activation. J. Immunol. 207, 1545–1554 (2021).
Kapur, R., Einarsdottir, H. K. & Vidarsson, G. IgG-effector functions: ‘the good, the bad and the ugly’. Immunol. Lett. 160, 139–144 (2014).
Shields, R. L. et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277, 26733–26740 (2002).
Shibata-Koyama, M. et al. Nonfucosylated rituximab potentiates human neutrophil phagocytosis through its high binding for FcγRIIIb and MHC class II expression on the phagocytotic neutrophils. Exp. Hematol. 37, 309–321 (2009).
Beck, A. & Reichert, J. M. Marketing approval of mogamulizumab: a triumph for glyco-engineering. MAbs 4, 419–425 (2012).
Dekkers, G. et al. Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of Fc-receptor- and complement-mediated-effector activities. Front. Immunol. 8, 877 (2017).
Vergroesen, R. D. et al. B-cell receptor sequencing of anti-citrullinated protein antibody (ACPA) IgG-expressing B cells indicates a selective advantage for the introduction of N-glycosylation sites during somatic hypermutation. Ann. Rheum. Dis. 77, 956–958 (2018).
Trbojevic-Akmacic, I., Abdel-Mohsen, M., Falck, D. & Rapp, E. Editorial: immunoglobulin glycosylation analysis: state-of-the-art methods and applications in immunology. Front. Immunol. 13, 923393 (2022).
Bondt, A. et al. ACPA IgG galactosylation associates with disease activity in pregnant patients with rheumatoid arthritis. Ann. Rheum. Dis. 77, 1130–1136 (2018).
Larsen, M. D. et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science 371, eabc8378 (2021).
Kapur, R. et al. A prominent lack of IgG1–Fc fucosylation of platelet alloantibodies in pregnancy. Blood 123, 471–480 (2014).
Bartsch, Y. C. et al. IgG Fc sialylation is regulated during the germinal center reaction following immunization with different adjuvants. J. Allergy Clin. Immunol. 146, 652–666 e611 (2020).
Wojcik, I. et al. A functional spleen contributes to afucosylated IgG in humans. Sci. Rep. 11, 24045 (2021).
Cao, Y. et al. Cytokines in the immune microenvironment change the glycosylation of IgG by regulating intracellular glycosyltransferases. Front. Immunol. 12, 724379 (2021).
Larsen, M. D. et al. Afucosylated Plasmodium falciparum-specific IgG is induced by infection but not by subunit vaccination. Nat. Commun. 12, 5838 (2021).
Buhre, J. S. et al. mRNA vaccines against SARS-CoV-2 induce comparably low long-term IgG Fc galactosylation and sialylation levels but increasing long-term IgG4 responses compared to an adenovirus-based vaccine. Front. Immunol. 13, 1020844 (2023).
Pongracz, T. et al. Immunoglobulin G1 Fc glycosylation as an early hallmark of severe COVID-19. EBioMedicine 78, 103957 (2022).
Kemna, M. J. et al. Galactosylation and sialylation levels of IgG predict relapse in patients with PR3–ANCA-associated vasculitis. EBioMedicine 17, 108–118 (2017).
Wang, J. et al. Fc-glycosylation of IgG1 is modulated by B-cell stimuli. Mol. Cell. Proteom. 10, 004655 (2011).
Ercan, A. et al. Estrogens regulate glycosylation of IgG in women and men. JCI Insight 2, e89703 (2017).
Washburn, N. et al. Controlled tetra-Fc sialylation of IVIg results in a drug candidate with consistent enhanced anti-inflammatory activity. Proc. Natl Acad. Sci. USA 112, E1297–E1306 (2015).
Falck, D., Jansen, B. C., de Haan, N. & Wuhrer, M. High-throughput analysis of IgG Fc glycopeptides by LC–MS. Methods Mol. Biol. 1503, 31–47 (2017).
Wuhrer, M. et al. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics 7, 4070–4081 (2007).
Selman, M. H. et al. Fc specific IgG glycosylation profiling by robust nano-reverse phase HPLC–MS using a sheath-flow ESI sprayer interface. J. Proteom. 75, 1318–1329 (2012).
Kammeijer, G. S. et al. Dopant enriched nitrogen gas combined with sheathless capillary electrophoresis-electrospray ionization-mass spectrometry for improved sensitivity and repeatability in glycopeptide analysis. Anal. Chem. 88, 5849–5856 (2016).
Falck, D. et al. Glycoforms of immunoglobulin G based biopharmaceuticals are differentially cleaved by trypsin due to the glycoform influence on higher-order structure. J. Proteome Res. 14, 4019–4028 (2015).
Simurina, M. et al. Glycosylation of immunoglobulin G associates with clinical features of inflammatory bowel diseases. Gastroenterology 154, e1310 (2018).
Jansen, B. C. et al. LaCyTools: a targeted liquid chromatography–mass spectrometry data processing package for relative quantitation of glycopeptides. J. Proteome Res. 15, 2198–2210 (2016).
Falck, D. et al. Glycoform-resolved pharmacokinetic studies in a rat model employing glycoengineered variants of a therapeutic monoclonal antibody. MAbs 13, 1865596 (2021).
Scherer, H. U. et al. Immunoglobulin 1 (IgG1) Fc-glycosylation profiling of anti-citrullinated peptide antibodies from human serum. Proteom. Clin. Appl. 3, 106–115 (2009).
Rombouts, Y. et al. Anti-citrullinated protein antibodies acquire a pro-inflammatory Fc glycosylation phenotype prior to the onset of rheumatoid arthritis. Ann. Rheum. Dis. 74, 234–241 (2015).
Wuhrer, M. et al. Skewed Fc glycosylation profiles of anti-proteinase 3 immunoglobulin G1 autoantibodies from granulomatosis with polyangiitis patients show low levels of bisection, galactosylation, and sialylation. J. Proteome Res. 14, 1657–1665 (2015).
Sonneveld, M. E. et al. Patients with IgG1-anti-red blood cell autoantibodies show aberrant Fc-glycosylation. Sci. Rep. 7, 8187 (2017).
Kapur, R. et al. Low anti-RhD IgG-Fc-fucosylation in pregnancy: a new variable predicting severity in haemolytic disease of the fetus and newborn. Br. J. Haematol. 166, 936–945 (2014).
Sonneveld, M. E. et al. Antigen specificity determines anti-red blood cell IgG-Fc alloantibody glycosylation and thereby severity of haemolytic disease of the fetus and newborn. Br. J. Haematol. 176, 651–660 (2017).
Sonneveld, M. E. et al. Fc-glycosylation in human IgG1 and IgG3 is similar for both total and anti-red-blood cell anti-K antibodies. Front. Immunol. 9, 129 (2018).
Siekman, S. L. et al. The IgG glycome of SARS-CoV-2 infected individuals reflects disease course and severity. Front. Immunol. 13, 993354 (2022).
Ackerman, M. E. et al. Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J. Clin. Invest. 123, 2183–2192 (2013).
Lu, L. L. et al. Antibody Fc glycosylation discriminates between latent and active tuberculosis. J. Infect. Dis. 222, 2093–2102 (2020).
Van Coillie, J. et al. The BNT162b2 mRNA SARS-CoV-2 vaccine induces transient afucosylated IgG1 in naive but not in antigen-experienced vaccinees. EBioMedicine 87, 104408 (2023).
van Osch, T. L. J. et al. Altered Fc glycosylation of anti-HLA alloantibodies in hemato-oncological patients receiving platelet transfusions. J. Thromb. Haemost. 20, 3011–3025 (2022).
Bharadwaj, P. et al. Afucosylation of HLA-specific IgG1 as a potential predictor of antibody pathogenicity in kidney transplantation. Cell Rep. Med. 3, 100818 (2022).
Sokolova, M. V. et al. Antibodies against citrullinated proteins of IgA isotype are associated with progression to rheumatoid arthritis in individuals at-risk. RMD Open. 9, e002705 (2023).
Lai, K. N. et al. IgA nephropathy. Nat. Rev. Dis. Prim. 2, 16001 (2016).
Steffen, U. et al. IgA subclasses have different effector functions associated with distinct glycosylation profiles. Nat. Commun. 11, 120 (2020).
Momcilovic, A. et al. Simultaneous immunoglobulin A and G glycopeptide profiling for high-throughput applications. Anal. Chem. 92, 4518–4526 (2020).
Chandler, K. B. et al. Multi-isotype glycoproteomic characterization of serum antibody heavy chains reveals isotype- and subclass-specific N-glycosylation profiles. Mol. Cell. Proteom. 18, 686–703 (2019).
Overall, M. L., Marzuki, S. & Hertzog, P. J. Comparison of different ELISAs for the detection of monoclonal antibodies to human interferon-α. Implications for antibody screening. J. Immunol. Methods 119, 27–33 (1989).
Yuan, S. et al. Changes in anti-thyroglobulin IgG glycosylation patterns in Hashimoto’s thyroiditis patients. J. Clin. Endocrinol. Metab. 100, 717–724 (2015).
Sahoo, A. et al. A clade C HIV-1 vaccine protects against heterologous SHIV infection by modulating IgG glycosylation and T helper response in macaques. Sci. Immunol. 7, eabl4102 (2022).
Aoyama, M. et al. Effects of terminal galactose residues in mannose α1-6 arm of Fc-glycan on the effector functions of therapeutic monoclonal antibodies. MAbs 11, 826–836 (2019).
Scherer, H. U. et al. Glycan profiling of anti-citrullinated protein antibodies isolated from human serum and synovial fluid. Arthritis Rheum. 62, 1620–1629 (2010).
Amez Martin, M., Wuhrer, M. & Falck, D. Serum and plasma immunoglobulin G Fc N-glycosylation is stable during Storage. J. Proteome Res. 20, 2935–2941 (2021).
Amez-Martin, M., Wuhrer, M. & Falck, D. Immunoglobulin G glycoprofiles are unaffected by common bottom-up sample processing. J. Proteome Res. 19, 4158–4162 (2020).
de Haan, N. et al. Developments and perspectives in high-throughput protein glycomics: enabling the analysis of thousands of samples. Glycobiology 32, 651–663 (2022).
Thermo Fisher Scientific Inc. Chromatography Troubleshooting Guides—Liquid Chromatography https://www.thermofisher.com/nl/en/home/industrial/chromatography/chromatography-learning-center/chromatography-consumables-resources/chromatography-troubleshooting-guides/chromatography-troubleshooting-guides-liquid-chromatography.html (2006).
Pongracz, T., Vidarsson, G. & Wuhrer, M. Antibody glycosylation in COVID-19. Glycoconj. J. 39, 335–344 (2022).
Vattepu, R., Sneed, S. L. & Anthony, R. M. Sialylation as an important regulator of antibody function. Front. Immunol. 13, 818736 (2022).
Bartsch, Y. C. et al. Sialylated autoantigen-reactive IgG antibodies attenuate disease development in autoimmune mouse models of lupus nephritis and rheumatoid arthritis. Front. Immunol. 9, 1183 (2018).
Sustic, T. et al. Immunoassay for quantification of antigen-specific IgG fucosylation. EBioMedicine 81, 104109 (2022).
Acknowledgements
We acknowledge support from the European Union (European Research Council Synergy, GlycanSwitch, Project 101071386) and the Dutch Research Council in the framework of the Domain Science (ENW) public–private partnership (PPP) Fund for the top sectors (Proteoform-resolved pharmacokinetics of biopharmaceuticals, project no. 019.012), with co-funding through the PPP Allowance made available by Health–Holland, Top Sector Life Sciences & Health. We thank M. Ceschi for the creating of the GlYcoLISA logo (included in Fig. 2).
Author information
Authors and Affiliations
Contributions
D.F. wrote and M.W. corrected the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Yusuke Mimura and the other, anonymous, reviewer(s) for their contribution to the peer review process of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references in the development of the protocol
Larsen, M. D. et al. Science 371, eabc8378 (2021): https://doi.org/10.1126/science.abc8378
Larsen, M. D. et al. Nat. Commun. 12, 5838 (2021): https://doi.org/10.1038/s41467-021-26118-w
Jansen, B. C. et al. J. Proteome Res. 15, 2198–2210 (2016): https://doi.org/10.1021/acs.jproteome.6b00171
Extended data
Supplementary information
Source data
Source Data Fig. 5
Relative abundances of glycans and glycosylation traits underlying Fig. 5.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Falck, D., Wuhrer, M. GlYcoLISA: antigen-specific and subclass-specific IgG Fc glycosylation analysis based on an immunosorbent assay with an LC–MS readout. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-00963-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-00963-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.