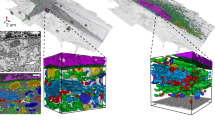
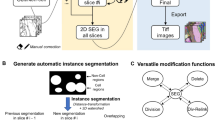
Abstract
Volume electron microscopy is the method of choice for the in situ interrogation of cellular ultrastructure at the nanometer scale, and with the increase in large raw image datasets generated, improving computational strategies for image segmentation and spatial analysis is necessary. Here we describe a practical and annotation-efficient pipeline for organelle-specific segmentation, spatial analysis and visualization of large volume electron microscopy datasets using freely available, user-friendly software tools that can be run on a single standard workstation. The procedures are aimed at researchers in the life sciences with modest computational expertise, who use volume electron microscopy and need to generate three-dimensional (3D) segmentation labels for different types of cell organelles while minimizing manual annotation efforts, to analyze the spatial interactions between organelle instances and to visualize the 3D segmentation results. We provide detailed guidelines for choosing well-suited segmentation tools for specific cell organelles, and to bridge compatibility issues between freely available open-source tools, we distribute the critical steps as easily installable Album solutions for deep learning segmentation, spatial analysis and 3D rendering. Our detailed description can serve as a reference for similar projects requiring particular strategies for single- or multiple-organelle analysis, which can be achieved with computational resources commonly available to single-user setups.
Key points
-
This protocol provides a pipeline for analyzing volume electron microscopy datasets covering the preparation of raw data, the segmentation of specific organelles, their spatial analysis and three-dimensional visualization of the segmentation maps.
-
The protocol demonstrates the use of tools such as Microscopy Image Browser, ilastik, Labkit and Album, which facilitates the installation of Python-based software (CSBDeep, CellSketch, StarDist, Blender and Jupyter notebooks).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout













Similar content being viewed by others
Data availability
All raw datasets at their original resolution are available at OpenOrganelle (https://openorganelle.janelia.org/). Demo data of raw data, segmentation masks, deep learning training data and spatial analysis are available at https://zenodo.org/record/8114392. Further information on the demo data can be found in the Supplementary Information.
Code availability
The code for Album is available at https://gitlab.com/album-app/album. The code for the Album solutions is available at https://github.com/betaseg/solutions. The code for the CellSketch viewer is available at https://github.com/betaseg/cellsketch. The Jupyter Notebooks for demo workflows can be found at https://github.com/betaseg/protocol-notebooks.
References
Peddie, C. J. & Collinson, L. M. Exploring the third dimension: volume electron microscopy comes of age. Micron 61, 9–19 (2014).
Peddie, C. J. et al. Volume electron microscopy. Nat. Rev. Methods Prim. 2, 51 (2022).
Hua, Y., Laserstein, P. & Helmstaedter, M. Large-volume en-bloc staining for electron microscopy-based connectomics. Nat. Commun. 6, 7923 (2015).
Kievits, A. J., Lane, R., Carroll, E. C. & Hoogenboom, J. P. How innovations in methodology offer new prospects for volume electron microscopy. J. Microsc. 287, 114–137 (2022).
Graham, B. J. et al. High-throughput transmission electron microscopy with automated serial sectioning. Preprint at bioRxiv https://doi.org/10.1101/657346 (2019).
Yin, W. et al. A petascale automated imaging pipeline for mapping neuronal circuits with high-throughput transmission electron microscopy. Nat. Commun. 11, 4949 (2020).
Phelps, J. S. et al. Reconstruction of motor control circuits in adult Drosophila using automated transmission electron microscopy. Cell 184, 759–774.e18 (2021).
Xu, C. S. et al. Enhanced FIB-SEM systems for large-volume 3D imaging. eLife 6, e25916 (2017).
Motta, A. et al. Dense connectomic reconstruction in layer 4 of the somatosensory cortex. Science 366, eaay3134 (2019).
Scheffer, L. K. et al. A connectome and analysis of the adult Drosophila central brain. eLife 9, e57443 (2020).
Müller, A. et al. 3D FIB-SEM reconstruction of microtubule-organelle interaction in whole primary mouse β. cells J. Cell Biol. 220, e202010039 (2021).
Parlakgül, G. et al. Regulation of liver subcellular architecture controls metabolic homeostasis. Nature 603, 736–742 (2022).
Sheu, S.-H. et al. A serotonergic axon-cilium synapse drives nuclear signaling to alter chromatin accessibility. Cell 185, 3390–3407.e18 (2022).
Weigel, A. V. et al. ER-to-Golgi protein delivery through an interwoven, tubular network extending from ER. Cell 184, 2412–2429.e16 (2021).
Uwizeye, C. et al. Morphological bases of phytoplankton energy management and physiological responses unveiled by 3D subcellular imaging. Nat. Commun. 12, 1049 (2021).
Musser, J. M. et al. Profiling cellular diversity in sponges informs animal cell type and nervous system evolution. Science 374, 717–723 (2021).
Bharathan, N. K. et al. Architecture and dynamics of a desmosome–endoplasmic reticulum complex. Nat. Cell Biol. 25, 823–835 (2023).
Malong, L. et al. Characterization of the structure and control of the blood-nerve barrier identifies avenues for therapeutic delivery. Dev. Cell 58, 174–191.e8 (2023).
Cortese, M. et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe 28, 853–866.e5 (2020).
Vergara, H. M. et al. Whole-body integration of gene expression and single-cell morphology. Cell 184, 4819–4837.e22 (2021).
Iudin, A., Korir, P. K., Salavert-Torres, J., Kleywegt, G. J. & Patwardhan, A. EMPIAR: a public archive for raw electron microscopy image data. Nat. Methods 13, 387–388 (2016).
Conrad, R. & Narayan, K. CEM500K, a large-scale heterogeneous unlabeled cellular electron microscopy image dataset for deep learning. eLife 10, e65894 (2021).
Xu, C. S. et al. An open-access volume electron microscopy atlas of whole cells and tissues. Nature 599, 147–151 (2021).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
Noske, A. B., Costin, A. J., Morgan, G. P. & Marsh, B. J. Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. J. Struct. Biol. 161, 298–313 (2008).
Wu, Y. et al. Contacts between the endoplasmic reticulum and other membranes in neurons. Proc. Natl Acad. Sci. USA 114, E4859–E4867 (2017).
Kaynig, V. et al. Large-scale automatic reconstruction of neuronal processes from electron microscopy images. Med. Image Anal. 22, 77–88 (2015).
Dorkenwald, S. et al. Automated synaptic connectivity inference for volume electron microscopy. Nat. Methods 14, 435–442 (2017).
Buhmann, J. et al. Automatic detection of synaptic partners in a whole-brain Drosophila electron microscopy data set. Nat. Methods 18, 771–774 (2021).
Heinrich, L. et al. Whole-cell organelle segmentation in volume electron microscopy. Nature 599, 141–146 (2021).
Spiers, H. et al. Deep learning for automatic segmentation of the nuclear envelope in electron microscopy data, trained with volunteer segmentations. Traffic 22, 240–253 (2021).
Gallusser, B. et al. Deep neural network automated segmentation of cellular structures in volume electron microscopy. J. Cell Biol. 222, e202208005 (2022).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Falk, T. et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat. Methods 16, 67–70 (2019).
Belevich, I., Joensuu, M., Kumar, D., Vihinen, H. & Jokitalo, E. Microscopy image browser: a platform for segmentation and analysis of multidimensional datasets. PLoS Biol. 14, e1002340 (2016).
Tu, Z. & Bai, X. Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 32, 1744–1757 (2010).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Kreshuk, A. & Zhang, C. in Computer Optimized Microscopy: Methods and Protocols (eds Rebollo, E. & Bosch, M.) 449–463 (Springer, 2019).
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Weigert, M., Schmidt, U., Haase, R., Sugawara, K. & Myers, G. Star-convex polyhedra for 3D object detection and segmentation in microscopy. In 2020 IEEE Winter Conference on Applications of Computer Vision (WACV) https://doi.org/10.1109/WACV45572.2020.9093435 (2020).
Helmstaedter, M., Briggman, K. L. & Denk, W. High-accuracy neurite reconstruction for high-throughput neuroanatomy. Nat. Neurosci. 14, 1081–1088 (2011).
Arzt, M. et al. LABKIT: labeling and segmentation toolkit for big image data. Front. Comput. Sci. https://doi.org/10.3389/fcomp.2022.777728 (2022).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Bolte, S. & Cordelières, F. P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006).
Shrestha, N. et al. Integration of ER protein quality control mechanisms defines β-cell function and ER architecture. J. Clin. Invest. https://doi.org/10.1172/JCI163584 (2022).
Schmid, B. et al. 3Dscript: animating 3D/4D microscopy data using a natural-language-based syntax. Nat. Methods 16, 278–280 (2019).
Albrecht, J. P., Schmidt, D. & Harrington, K. Album: a framework for scientific data processing with software solutions of heterogeneous tools. Preprint at https://doi.org/10.48550/arXiv.2110.00601 (2021).
Conrad, R. & Narayan, K. Instance segmentation of mitochondria in electron microscopy images with a generalist deep learning model trained on a diverse dataset. Cell Syst. 14, 58–71.e5 (2023).
von Chamier, L. et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun. 12, 2276 (2021).
Ouyang, W. et al. BioImage Model Zoo: a community-driven resource for accessible deep learning in bioimage analysis. Preprint at bioRxiv https://doi.org/10.1101/2022.06.07.495102 (2022).
Weber, B. et al. Automated tracing of microtubules in electron tomograms of plastic embedded samples of Caenorhabditis elegans embryos. J. Struct. Biol. 178, 129–138 (2012).
Eckstein, N., Buhmann, J., Cook, M. & Funke, J. Microtubule tracking in electron microscopy volumes. In Medical Image Computing and Computer Assisted Intervention (MICCAI) Part V 99–108 (Springer, 2020).
Kaltdorf, K. V. et al. Automated classification of synaptic vesicles in electron tomograms of C. elegans using machine learning. PLoS ONE 13, e0205348 (2018).
Haberl, M. G. et al. CDeep3M—plug-and-play cloud-based deep learning for image segmentation. Nat. Methods 15, 677–680 (2018).
Koranne, S. in Handbook of Open Source Tools (ed. Koranne, S.) 191–200 (Springer, 2011).
Saalfeld, S. et al. saalfeldlab/n5: n5-2.5.1 https://doi.org/10.5281/zenodo.6578232 (2022).
Miles, A. et al. zarr-developers/zarr-python: v2.4.0 https://doi.org/10.5281/zenodo.3773450 (2020).
Luengo, I. et al. SuRVoS: super-region volume segmentation workbench. J. Struct. Biol. 198, 43–53 (2017).
Pennington, A. et al. SuRVoS 2: accelerating annotation and segmentation for large volumetric bioimage workflows across modalities and scales. Front. Cell Dev. Biol. 10, 842342 (2022).
Belevich, I. & Jokitalo, E. DeepMIB: user-friendly and open-source software for training of deep learning network for biological image segmentation. PLoS Comput. Biol. 17, e1008374 (2021).
Hennies, J. et al. CebraEM: a practical workflow to segment cellular organelles in volume SEM datasets using a transferable CNN-based membrane prediction. Preprint at bioRxiv https://doi.org/10.1101/2023.04.06.535829 (2023).
Smith, P. et al. Online citizen science with the Zooniverse for analysis of biological volumetric data. Histochem. Cell Biol. https://doi.org/10.1007/s00418-023-02204-6 (2023).
Jorstad, A. et al. NeuroMorph: a toolset for the morphometric analysis and visualization of 3D models derived from electron microscopy image stacks. Neuroinformatics 13, 83–92 (2015).
Jorstad, A., Blanc, J. & Knott, G. NeuroMorph: a software toolset for 3D analysis of neurite morphology and connectivity. Front. Neuroanat. 12, 59 (2018).
Troidl, J. et al. Barrio: customizable spatial neighborhood analysis and comparison for nanoscale brain structures. Comput. Graph. Forum 41, 183–194 (2022).
Schroff, F., Criminisi, A. & Zisserman, A. in Procedings of the British Machine Vision Conference 2008 54.1–54.10 (British Machine Vision Association, 2008).
Arganda-Carreras, I. et al. Trainable weka segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33, 2424–2426 (2017).
Hallou, A., Yevick, H. G., Dumitrascu, B. & Uhlmann, V. Deep learning for bioimage analysis in developmental biology. Development 148, dev199616 (2021).
Shaga Devan, K., Kestler, H. A., Read, C. & Walther, P. Weighted average ensemble-based semantic segmentation in biological electron microscopy images. Histochem. Cell Biol. 158, 447–462 (2022).
Mandal, S. & Uhlmann, V. Splinedist: automated cell segmentation with spline curves. In 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI) https://doi.org/10.1109/ISBI48211.2021.9433928 (IEEE, 2021).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Sheridan, A. et al. Local shape descriptors for neuron segmentation. Nat. Methods 20, 295–303 (2023).
McDonald, K. L., O’Toole, E. T., Mastronarde, D. N. & McIntosh, J. R. Kinetochore microtubules in PTK cells. J. Cell Biol. 118, 369–383 (1992).
Marsh, B. J., Mastronarde, D. N., Buttle, K. F., Howell, K. E. & McIntosh, J. R. Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc. Natl Acad. Sci. USA 98, 2399–2406 (2001).
Rueden, C. T. et al. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinforma. 18, 529 (2017).
Pietzsch, T., Preibisch, S., Tomančák, P. & Saalfeld, S. ImgLib2—generic image processing in Java. Bioinformatics 28, 3009–3011 (2012).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Virtanen, P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
McKinney, W. Data structures for statistical computing in Python. In Proc. of the 9th Python in Science Conference. https://doi.org/10.25080/Majora-92bf1922-00a (2010).
Schmid, B., Schindelin, J., Cardona, A., Longair, M. & Heisenberg, M. A high-level 3D visualization API for Java and ImageJ. BMC Bioinforma. 11, 274 (2010).
Cardona, A. et al. TrakEM2 software for neural circuit reconstruction. PLoS ONE 7, e38011 (2012).
Hennies, J. et al. AMST: alignment to median smoothed template for focused ion beam scanning electron microscopy image stacks. Sci. Rep. 10, 2004 (2020).
Hanslovsky, P., Bogovic, J. A. & Saalfeld, S. Image-based correction of continuous and discontinuous non-planar axial distortion in serial section microscopy. Bioinformatics 33, 1379–1386 (2017).
Roels, J. et al. An interactive ImageJ plugin for semi-automated image denoising in electron microscopy. Nat. Commun. 11, 771 (2020).
Krull, A., Buchholz, T.-O. & Jug, F. Noise2Void—learning denoising from single noisy images. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2129–2137 (2019).
Perez, A. J. et al. A workflow for the automatic segmentation of organelles in electron microscopy image stacks. Front. Neuroanat. 8, 126 (2014).
Hoffman, D. P. et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357 (2020).
Müller, A. et al. Structure, interaction, and nervous connectivity of beta cell primary cilia. Preprint at bioRxriv https://doi.org/10.1101/2023.12.01.568979 (2024).
Park, G. et al. Amira annotation protocol. protocols.io https://www.protocols.io/view/amira-annotation-protocol-b834ryqw (2022).
Acknowledgements
We thank K. Pfriem for administrative assistance. We thank members of the PLID for valuable feedback. We thank S. Pang and C. Shan Xu (Yale University) as well as H. F. Hess (Janelia Research Campus) for FIB–SEM. We thank S. Kretschmar and T. Kurth from the Center for Molecular and Cellular Bioengineering Dresden (CMCB) for initial sample preparation. We thank all further authors of the original Journal of Cell Biology publication, J. Verner D’Costa, C. Münster (both PLID), and F. Jug (Human Technopole) for their support. This work was supported by the Electron Microscopy and Histology Facility, a Core Facility of the CMCB Technology Platform at TU Dresden. We thank B. Busselman (Universtiy of South Dakota) for testing Album installation. We thank the EM facility of the Max Planck Institute of Molecular Cell Biology and Genetics for their services. This work was supported with funds to M.S. from the German Center for Diabetes Research by the German Ministry for Education and Research (BMBF) and from the Innovative Medicines Initiative 2 Joint Undertaking under grant agreement no. 115881 (RHAPSODY) and 115797 (INNODIA). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program and the European Federation of Pharmaceutical Industries and Associations (EFPIA). This work is further supported by the Swiss State Secretariat for Education‚ Research and Innovation under contract no. 16.0097-2. A.M. was the recipient of a MeDDrive grant from the Carl Gustav Carus Faculty of Medicine at TU Dresden. M.W. was supported by the ELISIR program of the École Polytechnique Fédérale de Lausanne School of Life Sciences and by generous funding from CARIGEST SA. D.S. and L.R. were funded by Helmholtz Imaging, a platform of the Helmholtz Information & Data Science Incubator.
Author information
Authors and Affiliations
Contributions
A.M., D.S., M.S. and M.W. wrote the manuscript. D.S., J.P.A. and M.W. wrote the workflow implementations. L.R., M.O., G.F. and L.E.G.G. tested the workflows and provided feedback on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Kedar Narayan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Müller, A. et al. J. Cell Biol. 220, e202010039 (2021): https://doi.org/10.1083/jcb.202010039
Supplementary information
Supplementary Information
Supplementary workflows and Table 1.
Supplementary Video 1
Album/CSBDeep training. How to use the Album GUI to install and execute the CSBDeep Album solution that trains a U-Net for semantic segmentation of Golgi from FIB–SEM volumes. This includes visualization of the loss and intermediate results via tensorboard.
Supplementary Video 2
Album/StarDist training. How to use the Album GUI to install and execute the StarDist Album solution that trains a StarDist network for instance segmentation of SGs from FIB–SEM volumes. It additionally shows how to set and change training parameters and how to visualize the loss and intermediate results via tensorboard.
Supplementary Video 3
Album/CellSketch project creation. How to install the protocol solutions catalog and how to create a CellSketch project via the Album GUI. In the video, several cell component annotations are added to the project.
Supplementary Video 4
Album/CellSketch spatial analysis. How to run the automated spatial analysis routine on an existing CellSketch project via the Album GUI. We also demonstrate how to visualize the analysis results in BigDataViewer and as Jupyter Notebook plots via Album solutions.
Supplementary Video 5
Album/CellSketch 3D rendering with Blender. How to convert pixel-based datasets from an existing CellSketch project into meshes via the Album GUI. We demonstrate how to visualize these meshes in VTK and how to automatically create a Blender scene including these meshes. Finally, the scene is rendered in Blender.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Müller, A., Schmidt, D., Albrecht, J.P. et al. Modular segmentation, spatial analysis and visualization of volume electron microscopy datasets. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-00957-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-00957-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.