Abstract
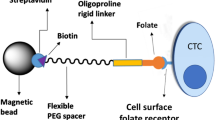
Identification and characterization of circulating tumor cells (CTCs) from blood samples of patients with cancer can help monitor parameters such as disease stage, disease progression and therapeutic efficiency. However, the sensitivity and specificity of current multivalent approaches used for CTC capture is limited by the lack of control over the ligands’ position. In this Protocol Update, we describe DNA-tetrahedral frameworks anchored with aptamers that can be configured with user-defined spatial arrangements and stoichiometries. The modified tetrahedral DNA frameworks, termed ‘n-simplexes’, can be used as probes to specifically target receptor−ligand interactions on the cell membrane. Here, we describe the synthesis and use of n-simplexes that target the epithelial cell adhesion molecule expressed on the surface of CTCs. The characterization of the n-simplexes includes measuring the binding affinity to the membrane receptors as a result of the spatial arrangement and stoichiometry of the aptamers. We further detail the capture of CTCs from patient blood samples. The procedure for the preparation and characterization of n-simplexes requires 11.5 h, CTC capture from clinical samples and data processing requires ~5 h per six samples and the downstream analysis of captured cells typically requires 5.5 h. The protocol is suitable for users with basic expertise in molecular biology and handling of clinical samples.
Key points
-
The procedure describes the design of targeted probes via orthogonal anchoring of aptamers, with varying stoichiometries, to the vertices of tetrahedral DNA frameworks. The frameworks, synthesized by using a single annealing step, are then characterized by using native polyacrylamide gel electrophoresis, atomic force microscopy, single-molecule fluorescence and transmission electron microscopy.
-
The targeted tetrahedral DNA frameworks improve CTC capture efficiency when compared with alternative nucleic acid–based probes in whole-blood samples with the supernatant removed via centrifugation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The main data discussed in this protocol are available in the supporting primary research paper29. The raw datasets are too large to be publicly shared but are available for research purposes from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Alix-Panabières, C. & Pantel, K. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 59, 110–118 (2013).
Ma, N. & Jeffrey, S. S. Deciphering cancer clues from blood. Science 367, 1424–1425 (2020).
Kelley, S. O. & Pantel, K. A new era in liquid biopsy: from genotype to phenotype. Clin. Chem. 66, 89–96 (2020).
Ozkumur, E. et al. Inertial focusing for tumor antigen–dependent and –independent sorting of rare circulating tumor cells. Sci. Transl. Med. 5, 179ra147 (2013).
Marquette, C. H. et al. Circulating tumour cells as a potential biomarker for lung cancer screening: a prospective cohort study. Lancet Respir. Med. 8, 709–716 (2020).
Steeg, P. S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 12, 895–904 (2006).
Qin, W. et al. Bioinspired DNA nanointerface with anisotropic aptamers for accurate capture of circulating tumor cells. Adv. Sci. (Weinh.) 7, 2000647 (2020).
Song, Y. et al. Bioinspired engineering of a multivalent aptamer-functionalized nanointerface to enhance the capture and release of circulating tumor cells. Angew. Chem. Int. Ed. Engl. 58, 2236–2240 (2019).
Englund, E. A. et al. Programmable multivalent display of receptor ligands using peptide nucleic acid nanoscaffolds. Nat. Commun. 3, 614 (2012).
Zhao, W. et al. Bioinspired multivalent DNA network for capture and release of cells. Proc. Natl Acad. Sci. USA 109, 19626–19631 (2012).
Sheng, W., Chen, T., Tan, W. & Fan, Z. H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano 7, 7067–7076 (2013).
Yoon, H. J. et al. Sensitive capture of circulating tumour cells by functionalized graphene oxide nanosheets. Nat. Nanotechnol. 8, 735–741 (2013).
Shen, Z., Wu, A. & Chen, X. Current detection technologies for circulating tumor cells. Chem. Soc. Rev. 46, 2038–2056 (2017).
Gomes de Castro, M. A. et al. Differential organization of tonic and chronic B cell antigen receptors in the plasma membrane. Nat. Commun. 10, 820 (2019).
Huppa, J. B. et al. TCR–peptide–MHC interactions in situ show accelerated kinetics and increased affinity. Nature 463, 963–967 (2010).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Li, M. et al. Nucleic acid tests for clinical translation. Chem. Rev. 121, 10469–10558 (2021).
Lin, M. et al. Programmable engineering of a biosensing interface with tetrahedral DNA nanostructures for ultrasensitive DNA detection. Angew. Chem. Int. Ed. Engl. 54, 2151–2155 (2015).
Liu, Q. et al. Valency-controlled framework nucleic acid signal amplifiers. Angew. Chem. Int. Ed. Engl. 57, 7131–7135 (2018).
Yin, F. et al. DNA-framework-based multidimensional molecular classifiers for cancer diagnosis. Nat. Nanotechnol. 18, 677–686 (2023).
Ye, D., Zuo, X. & Fan, C. DNA nanotechnology-enabled interfacial engineering for biosensor development. Annu. Rev. Anal. Chem. 11, 171–195 (2018).
Zhao, S. et al. A DNA origami-based aptamer nanoarray for potent and reversible anticoagulation in hemodialysis. Nat. Commun. 12, 358 (2021).
Wilner, O. I. & Willner, I. Functionalized DNA nanostructures. Chem. Rev. 112, 2528–2556 (2012).
Chandrasekaran, A. R., Anderson, N., Kizer, M., Halvorsen, K. & Wang, X. Beyond the fold: emerging biological applications of DNA origami. Chembiochem 17, 1081–1089 (2016).
Kacherovsky, N. et al. Traceless aptamer-mediated isolation of CD8+ T cells for chimeric antigen receptor T-cell therapy. Nat. Biomed. Eng. 3, 783–795 (2019).
Mao, M. et al. Modular DNA-origami-based nanoarrays enhance cell binding affinity through the “lock-and-key” interaction. J. Am. Chem. Soc. 145, 5447–5455 (2023).
Lin, M. et al. Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform. Nat. Protoc. 11, 1244–1263 (2016).
Li, M. et al. DNA framework-programmed cell capture via topology-engineered receptor–ligand interactions. J. Am. Chem. Soc. 141, 18910–18915 (2019).
Zhang, J. L. et al. DNA nanolithography enables a highly ordered recognition interface in a microfluidic chip for the efficient capture and release of circulating tumor cells. Angew. Chem. Int. Ed. Engl. 59, 14115–14119 (2020).
Chen, J. et al. Biospecific monolayer coating for multivalent capture of circulating tumor cells with high sensitivity. Adv. Funct. Mater. 29, 1808961 (2019).
Mitchell, N. et al. A DNA nanostructure for the functional assembly of chemical groups with tunable stoichiometry and defined nanoscale geometry. Angew. Chem. Int. Ed. Engl. 48, 525–527 (2008).
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2021YFF1200300 and 2020YFA0909000), the National Natural Science Foundation of China (22025404, T2188102 and 22174132), the Natural Science Foundation of Shanghai (23ZR1438700), the Shanghai Municipal Health Commission (2022JC027) and the Innovative Research Group of High-Level Local Universities in Shanghai (SHSMU-ZLCX20212602).
Author information
Authors and Affiliations
Contributions
M.Li and X.Z. supervised the projects. Y.C., M.Lin and D.Y. designed and conducted the experiments. Y.C., M.Lin and S.W. analyzed the data. Y.C. and M.Li wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Julian Alexander Tanner and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Related links
Key reference using this protocol
Li, M. et al. J. Am. Chem. Soc. 141, 18910–18915 (2019): https://doi.org/10.1021/jacs.9b11015
This protocol is an update to: Nat. Protoc. 11, 1244–1263 (2016): https://doi.org/10.1038/nprot.2016.071
Supplementary information
Supplementary Information
Supplementary Figs. 1–7
Supplementary Data 5
Unprocessed gels for Supplementary Fig. 5
Source data
Source Data Fig. 3
Unprocessed gels
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, Y., Lin, M., Ye, D. et al. Functionalized tetrahedral DNA frameworks for the capture of circulating tumor cells. Nat Protoc 19, 985–1014 (2024). https://doi.org/10.1038/s41596-023-00943-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00943-3
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.