Abstract
B cells generate antibodies that provide protection from infection, but also cause pathology in autoimmune and allergic conditions. Antigen-specific B cells can be detected by binding their surface antibody receptors with native antigens conjugated to fluorescent probes, a technique that has revealed substantial insight into B cell activation and function. This protocol describes the process of generating fluorescent antigen tetramer probes and delineates a process of enriching large samples based on antigen-specificity for high-resolution analyses of the antigen-specific B cell repertoire. Enrichment of tetramer-binding cells allows for detection of antigen-specific B cells as rare as 1 in 100 million cells, providing sufficient resolution to study naive B cells and IgE-expressing cells by flow cytometry. The generation of antigen tetramers involves antigen biotinylation, assessment of biotin:antigen ratio for optimal tetramer loading and polymerization around a streptavidin–fluorophore backbone. We also describe the construction of a control tetramer to exclude B cells binding to the tetramer backbone. We provide a framework to validate whether tetramer probes are detecting true antigen-specific B cells and discuss considerations for experimental design. This protocol can be performed by researchers trained in basic biomedical/immunological research techniques, using instrumentation commonly found in most laboratories. Constructing the antigen and control tetramers takes 9 h, though their specificity should be assessed before experimentation and may take weeks to months depending on the method of validation. Sample enrichment requires ~2 h but is generally time and cost neutral as fewer cells are run through the flow cytometer.
Key points
-
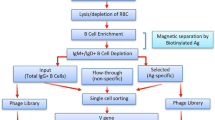
The protocol describes the design of antigen-specific tetramer probes for the enrichment and identification of rare antigen-specific B cells. The selected antigen is first biotinylated in a biotin:antigen ratio <1:1 and then assembled around a streptavidin fluorescent backbone.
-
Isolation of antigen-specific B cells relies on the accurate design of control tetramers that do not share any known cross-reactive epitopes with the target antigen.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are represented within the paper. Raw data files are available upon request. The corresponding authors have repositories of antigens used for antigen tetramer construction, which can be shared upon request. They are also willing to assist in construction of novel antigen tetramers, if contacted.
References
Young, C. & Brink, R. The unique biology of germinal center B cells. Immunity 54, 1652–1664 (2021).
Elsner, R. A. & Shlomchik, M. J. Germinal center and extrafollicular B cell responses in vaccination, immunity, and autoimmunity. Immunity 53, 1136–1150 (2020).
Sharonov, G. V., Serebrovskaya, E. O., Yuzhakova, D. V., Britanova, O. V. & Chudakov, D. M. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat. Rev. Immunol. 20, 294–307 (2020).
Cyster, J. G. & Allen, C. D. C. B cell responses: cell interaction dynamics and decisions. Cell 177, 524–540 (2019).
Lu, L. L., Suscovich, T. J., Fortune, S. M. & Alter, G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18, 46–61 (2017).
Tordesillas, L., Berin, M. C. & Sampson, H. A. Immunology of food allergy. Immunity 47, 32–50 (2017).
Bousquet, J. et al. Allergic rhinitis. Nat. Rev. Dis. Prim. 6, 95 (2020).
Weyand, C. M. & Goronzy, J. J. The immunology of rheumatoid arthritis. Nat. Immunol. 22, 10–18 (2020).
Koenig, J. F. et al. Memory generation and re-activation in food allergy. Immunotargets Ther. 10, 171–184 (2021).
Allman, D., Wilmore, J. R. & Gaudette, B. T. The continuing story of T-cell independent antibodies. Immunol. Rev. 288, 128–135 (2019).
Stavnezer, J., Guikema, J. E. J. & Schrader, C. E. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26, 261–292 (2008).
Pape, K. A., Taylor, J. J., Maul, R. W., Gearhart, P. J. & Jenkins, M. K. Different B cell populations mediate early and late memory during an endogenous immune response. Science 331, 1203–1207 (2011).
Boonyaratanakornkit, J. & Taylor, J. J. Techniques to study antigen-specific B cell responses. Front. Immunol. 10, 1694 (2019).
Moody, M. A. & Haynes, B. F. Antigen-specific B cell detection reagents: use and quality control. Cytom. A 73, 1086–1092 (2008).
Kodituwakku, A. P., Jessup, C., Zola, H. & Roberton, D. M. Isolation of antigen-specific B cells. Immunol. Cell Biol. 81, 163–170 (2003).
Franz, B., May, K. F., Dranoff, G. & Wucherpfennig, K. Ex vivo characterization and isolation of rare memory B cells with antigen tetramers. Blood 118, 348–357 (2011).
Taylor, J. J., Pape, K. A., Steach, H. R. & Jenkins, M. K. Apoptosis and antigen affinity limit effector cell differentiation of a single naive B cell. Science 347, 784–787 (2015).
Taylor, J. J., Pape, K. A. & Jenkins, M. K. A germinal center-independent pathway generates unswitched memory B cells early in the primary response. J. Exp. Med. 209, 597–606 (2012).
Taylor, J. J. et al. Deletion and anergy of polyclonal B cells specific for ubiquitous membrane-bound self-antigen. J. Exp. Med. 209, 2065–2077 (2012).
Boonyaratanakornkit, J. et al. Protective antibodies against human parainfluenza virus type 3 infection. MAbs 13, 1912884 (2021).
Koenig, J. F. E. et al. A distinct phenotype of polarized memory B cell holds IgE memory. Preprint at bioRxiv https://doi.org/10.1101/2023.01.25.525495 (2023).
Steach, H. R. et al. Cross-reactivity with self-antigen tunes the functional potential of naive B cells specific for foreign antigens. J. Immunol. 204, 498–509 (2020).
Aranda, C. J. et al. IgG memory B cells expressing IL4R and FCER2 are associated with atopic diseases. Allergy https://doi.org/10.1111/all.15601 (2022).
Bancroft, T. et al. Detection and activation of HIV broadly neutralizing antibody precursor B cells using anti-idiotypes. J. Exp. Med. 216, 2331–2347 (2019).
Cabán, M. et al. Cross-protective antibodies against common endemic respiratory viruses. Nat. Commun. 14, 798 (2023).
Krishnamurty, A. T. et al. Somatically hypermutated plasmodium-specific IgM+ memory B cells are rapid, plastic, early responders upon malaria rechallenge. Immunity 45, 402–414 (2016).
Coelho, C. H. et al. A human monoclonal antibody blocks malaria transmission and defines a highly conserved neutralizing epitope on gametes. Nat. Commun. 12, 1750 (2021).
Taylor, J. J., Laudenbach, M., Tucker, A. M., Jenkins, M. K. & Pravetoni, M. Hapten-specific naïve B cells are biomarkers of vaccine efficacy against drugs of abuse. J. Immunol. Methods 405, 74–86 (2014).
Rodda, L. B. et al. Functional SARS-CoV-2-specific immune memory persists after mild COVID-19. Cell 184, 169–183.e17 (2021).
Pape, K. A. et al. High-affinity memory B cells induced by SARS-CoV-2 infection produce more plasmablasts and atypical memory B cells than those primed by mRNA vaccines. Cell Rep. 37, 109823 (2021).
Afkhami, S. et al. Respiratory mucosal delivery of next-generation COVID-19 vaccine provides robust protection against both ancestral and variant strains of SARS-CoV-2. Cell 185, 896–915.e19 (2022).
Rodda, L. B. et al. Imprinted SARS-CoV-2-specific memory lymphocytes define hybrid immunity. Cell 185, 1588–1601.e14 (2022).
Saletti, G., Çuburu, N., Yang, J. S., Dey, A. & Czerkinsky, C. Enzyme-linked immunospot assays for direct ex vivo measurement of vaccine-induced human humoral immune responses in blood. Nat. Protoc. 8, 1073–1087 (2013).
Czerkinsky, C. C., Nilsson, L. Å., Nygren, H., Ouchterlony, Ö. & Tarkowski, A. A solid-phase enzyme-linked immunospot (ELISPOT) assay for enumeration of specific antibody-secreting cells. J. Immunol. Methods 65, 109–121 (1983).
Blanchard-Rohner, G., Galli, G., Clutterbuck, E. A. & Pollard, A. J. Comparison of a limiting dilution assay and ELISpot for detection of memory B-cells before and after immunisation with a protein-polysaccharide conjugate vaccine in children. J. Immunol. Methods 358, 46–55 (2010).
Townsend, S. E., Goodnow, C. C. & Cornall, R. J. Single epitope multiple staining to detect ultralow frequency B cells. J. Immunol. Methods 249, 137–146 (2001).
Leyendeckers, H. et al. Frequent detection of thyroid peroxidase-specific IgG+ memory B cells in blood of patients with autoimmune thyroid disease. Eur. J. Immunol. 32, 3126–3132 (2002).
Leyendeckers, H. et al. Correlation analysis between frequencies of circulating antigen-specific IgG-bearing memory B cells and serum titers of antigen-specific IgG. Eur. J. Immunol. 29, 1406–1417 (1999).
Irsch, J. et al. Isolation and characterization of allergen-binding cells from normal and allergic donors. Immunotechnology 1, 115–125 (1995).
Wennhold, K. et al. Using antigen-specific B cells to combine antibody and T cell-based cancer immunotherapy. Cancer Immunol. Res. 5, 730–743 (2017).
Newman, J., Rice, J. S., Wang, C., Harris, S. L. & Diamond, B. Identification of an antigen-specific B cell population. J. Immunol. Methods 272, 177–187 (2003).
Martin, F., Oliver, A. M. & Kearney, J. F. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity 14, 617–629 (2001).
Leyendeckers, H. et al. Memory B cells specific for the NC16A domain of the 180 kDa bullous pemphigoid autoantigen can be detected in peripheral blood of bullous pemphigoid patients and induced in vitro to synthesize autoantibodies. J. Invest. Dermatol. 120, 372–378 (2003).
Rahe, M. C., Gustafson, K. L. & Murtaugh, M. P. B cell tetramer development for veterinary vaccinology. Viral Immunol. 31, 1–10 (2018).
Fitzpatrick, K. S. et al. Validation of ligand tetramers for the detection of antigen-specific lymphocytes. J. Immunol. 210, 1156–1165 (2023).
Wade-Vallance, A. K. & Allen, C. D. C. Intrinsic and extrinsic regulation of IgE B cell responses. Curr. Opin. Immunol. 72, 221–229 (2021).
Berendam, S. J. et al. Different adjuvanted pediatric HIV envelope vaccines induced distinct plasma antibody responses despite similar B cell receptor repertoires in infant rhesus macaques. PLoS ONE 16, e0256885 (2021).
Beckett, D., Kovaleva, E. & Schatz, P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 8, 921–929 (1999).
Spanier, J. A. et al. Efficient generation of monoclonal antibodies against peptide in the context of MHCII using magnetic enrichment. Nat. Commun. 7, 1–11 (2016).
Pape, K. A. et al. Naive B cells with high-avidity germline-encoded antigen receptors produce persistent IgM+ and transient IgG+ memory B cells. Immunity 48, 1135–1143.e4 (2018).
Yang, Z., Sullivan, B. M. & Allen, C. D. C. Fluorescent in vivo detection reveals that IgE+ B cells are restrained by an intrinsic cell fate predisposition. Immunity 36, 857–872 (2012).
Haniuda, K., Fukao, S., Kodama, T., Hasegawa, H. & Kitamura, D. Autonomous membrane IgE signaling prevents IgE-memory formation. Nat. Immunol. 17, 1109–1117 (2016).
Jimenez-Saiz, R. et al. Human BCR analysis of single-sorted, putative IgE+ memory B cells in food allergy. J. Allergy Clin. Immunol. 144, 336–339.e6 (2019).
Yang, Z. et al. Regulation of B cell fate by chronic activity of the IgE B cell receptor. eLife 5, 1–31 (2016).
Glass, D. R. et al. An integrated multi-omic single-cell atlas of human B cell identity. Immunity 53, 217–232.e5 (2020).
Hill, S. W., Kipp, D. E., Melchers, I., Frelinger, J. A. & Sercarz, E. E. Multiple H-2 and non-H-2 genes controlling the anti-lysozyme response: alternative gene constellations can lead to responsiveness. Eur. J. Immunol. 10, 384–391 (1980).
Haniuda, K. & Kitamura, D. Induced germinal center B cell culture system. Bio. Protoc. 9, e3163 (2019).
Tiller, T. et al. Efficient generation of monoclonal antibodies from single human B cells by single cell RT–PCR and expression vector cloning. J. Immunol. Methods 329, 112–124 (2008).
Mason, D. Y., Jones, M. & Goodnow, C. C. Development and follicular localization of tolerant B lymphocytes in lysozyme/anti-lysozyme lgM/lgD transgenic mice. Int. Immunol. 4, 163–175 (1992).
Acknowledgements
We thank J. SoRelle (University of Texas Southwestern) for carefully reviewing and providing suggestions on the manuscript. We thank M. K. Jenkins (University of Minnesota) for supporting the initial development of these protocols and for the inclusion of Extended Data Fig. 1g. We thank J. Carter and D. Galloway (Fred Hutchinson Cancer Center) for providing supernatant containing GST. We thank the McMaster Flow Cytometry Core, H. Liang and M. Subapanditha for access to flow cytometers and experimental support. We thank M. S. Miller (McMaster University) and M. Larche (McMaster University) for providing recombinant RBD. The laboratories of M.J. and J.F.E.K. are funded by the Schroeder Foundation, Food Allergy Canada, ALK-Abello A/S, the Canadian Allergy Asthma and Immunology Foundation, the Zych family and the Satov family. A.P. is funded by an Ontario Graduate Scholarship and The Eva Eugenia Lillian Cope Scholarship. D.P.-C. was funded by Universidad Politécnica de Madrid and Banco Santander with predoctoral and travel Programa Propio grants. J.T.-A. was funded by Severo Ochoa Program (Production of Plant and Human health-relevant proteins in Super-Green Biofactories: PCD-UPM/7/2022). The Centre for Plant Biotechnology and Genomics was granted ‘Severo Ochoa’ Distinctions of Excellence by the Spanish Ministry of Science and Innovation (SEV-2016-0672 and CEX2020-000999-S). J.J.T. has been funded by the National Institutes of Health (R01AI122912, R01AI158728, R01AI167009), The Hartwell Foundation, Vir Biotechnology, Fred Hutch Cancer Center and generous donors. J.B. was supported by a Fast Grants award, and J.B. and J.J.T. were supported by a Fred Hutchinson Cancer Center COVID pilot award.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.P., D.P.-C., J.J.T. and J.F.E.K. Methodology: A.P., D.P.-C., J.J.T. and J.F.E.K. Formal analysis: A.P., D.P.-C., J.B., J.J.T. and J.F.E.K. Investigation: A.P., D.P.-C., F.U., E.G., O.M.-D., A.F., T.D.W., R.T.G., J.T.-A., A.D.-P., J.B. and J.F.E.K. Resources: J.T.-A., A.D.-P. and S.W. Visualization: A.P., D.P.-C., J.J.T. and J.F.E.K. Funding acquisition: J.T.-A., A.D.-P., S.W., M.J., J.J.T. and J.F.E.K. Supervision: J.J.T. and J.F.E.K. Writing — original draft: A.P., D.P.-C., J.J.T. and J.F.E.K. Writing — review and editing: A.P., D.P.-C., F.U., J.B., M.J., J.J.T. and J.F.E.K.
Corresponding authors
Ethics declarations
Competing interests
J.F.E.K. and M.J. receive funding from ALK Abello A/S. M.J. is an advisor for ALK Abello A/S. All other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Michael Anthony Moody and Gabrielle Rizzuto for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Taylor, J. J. et al. J. Exp. Med. 209, 2065–2077 (2012): https://doi.org/10.1084/jem.20112272
Koenig, J. F. E. et al. Preprint at bioRxiv (2023): https://doi.org/10.1101/2023.01.25.525495
Taylor J. J. et al. Science 347, (2015): https://doi.org/10.1126/science.aaa1342
Cabán, M. et al. Nat. Commun. 14, 798 (2023): https://doi.org/10.1038/s41467-023-36459-3
Bancroft, T. et al. J. Exp. Med. 216, 2331–2347 (2019): https://doi.org/10.1084/jem.20190164
Extended data
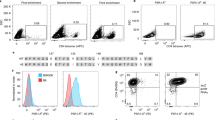
Extended Data Fig. 1 Antigen monomer and tetramer staining.
a) Representative plot of OVA-FITC monomer-stained samples from the mesenteric lymph nodes of mice sensitized with 4 gavages of OVA/CT. b) Isotype of OVA-FITC monomer-binding B cells from unimmunized mice. c) Representative plot of OVA-PE tetramer-stained samples from the spleen and mesenteric lymph nodes of unimmunized and OVA/Alum immunized mice. d) Comparison of SA-PE and OVA PE tetramer staining in unimmunized mice. e) Representative plot of control and antigen tetramer staining of spleen and mesenteric lymph nodes from unimmunized mice. f) Isotype of OVA PE tetramer-binding B cells from unimmunized mice. g) Demonstration of exclusion of backbone-binding B cells using a control tetramer. Comparison of unimmunized mice, mice immunized intraperitoneally with OVA/CFA, PE/CFA, and SA/CFA. H) RBD PE and RBD APC tetramer staining of anti-PE, anti-APC, anti-SA, and anti-RBD loaded compensation beads.
Extended Data Fig. 2 Antigen-specific B cell tracing by adoptive transfer of tetramer enriched cells.
a) Schematic of the experiment. b) Representative plots and quantification of control and OVA tetramer staining of spleen and mesenteric lymph nodes of recipient µMT mice 9 days after OVA/Alum immunization. c) Quantification of serum OVA-specific IgE and IgG1 antibodies by ELISA. d–f) Evaluation of phenotype of OVA-specific B-lineage cells in the enriched fraction of OVA/Alum-immunized µMT recipients. * p<0.05 by two-way ANOVA. Error bars represent SEM. Panel a created with BioRender.com.
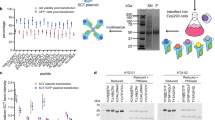
Extended Data Fig. 3 Examples of successful and failed blots to determine biotinylation ratio.
Sample blots of a) target biotinylation ratio, b) over-biotinylated antigen, c) under-biotinylated antigen.
Extended Data Fig. 4 Antigen tetramer enrichment from other tissues.
Evaluation of OVA tetramer binding in the unenriched, enriched and flow through fractions and evaluation of phenotype and isotype of OVA-specific B cells within the enriched fraction. a) OVA tetramer enrichment from unimmunized mice or mice sensitized with 4 gavages of OVA/CT. Small intestines from 5 mice were pooled prior to enrichment. b) OVA tetramer enrichment from unimmunized mice or mice immunized with an intraperitoneal injection of OVA/Alum. Bone marrow from the hind legs of one mouse is represented on each plot.
Extended Data Fig. 5 Example antigen tetramer reagents.
Enrichment of antigen tetramer-binding B cells from the spleen and mesenteric lymph nodes of unimmunized mice and those intraperitonally immunized with (a) Beta lactoglobulin (BLG), (b) Alt a 1, (c) Ara h 1, (d) SARS-CoV-2 spike RBD.
Extended Data Fig. 6 Additional information for Fig. 3.
a) Evaluation of phenotype of OVA-specific switched and IgE-expressing B cells from OVA/Alum immunized Verigem mice at 7 days post-immunization. b) Concatenated plot of intracellular IgE staining from 3 mice sensitized by 4 gavages of OVA/CT. Data was collected at day 6 after the last gavage. c) Extracellular IgG1 staining of OVA tetramer-binding B cells from unimmunized and OVA/Alum immunized mice. d) Intracellular IgG1 staining with and without blocking surface IgG1 with an unlabeled antibody.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Text and Methods.
Supplementary Software
Programmed Excel worksheet that performs calculations for this protocol.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Phelps, A., Pazos-Castro, D., Urselli, F. et al. Production and use of antigen tetramers to study antigen-specific B cells. Nat Protoc 19, 727–751 (2024). https://doi.org/10.1038/s41596-023-00930-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00930-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.