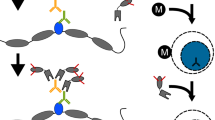
Abstract
Cleavage under targets and tagmentation (CUT&Tag) is an antibody-directed in situ chromatin profiling strategy that is rapidly replacing immune precipitation-based methods, such as chromatin immunoprecipitation–sequencing. The efficiency of the method enables chromatin profiling in single cells but is limited by the numbers of cells that can be profiled. Here, we describe a combinatorial barcoding strategy for CUT&Tag that harnesses a nanowell dispenser for simple, high-resolution, high-throughput, single-cell chromatin profiling. In this single-cell combinatorial indexing CUT&Tag (sciCUT&Tag) protocol, lightly cross-linked nuclei are bound to magnetic beads and incubated with primary and secondary antibodies in bulk and then arrayed in a 96-well plate for a first round of cellular indexing by antibody-directed Tn5 tagmentation. The sample is then repooled, mixed and arrayed across 5,184 nanowells at a density of 12–24 nuclei per well for a second round of cellular indexing during PCR amplification of the sequencing-ready library. This protocol can be completed in 1.5 days by a research technician, and we illustrate the optimized protocol by profiling histone modifications associated with developmental gene repression (H3K27me3) as well as transcriptional activation (H3K4me1-2-3) in human peripheral blood mononuclear cells and use single-nucleotide polymorphisms to facilitate collision removal. We have also used sciCUT&Tag for simultaneous profiling of multiple chromatin epitopes in single cells. The reduced cost, improved resolution and scalability of sciCUT&Tag make it an attractive platform to profile chromatin features in single cells.
Key points
-
This protocol describes a method for the single-cell profiling of epigenetic marks.
-
Compared with other single-cell chromatin profiling methods, single-cell combinatorial indexing cleavage under targets and tagmentation can profile a greater number of cells while maintaining the optimal reaction chemistry for efficient PCR amplification of tagmented chromatin, thereby increasing the number of unique reads recovered per cell relative to droplet-based approaches.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The primary sequencing data from the barnyard experiment, mixing human and mouse cell lines, as well as single-donor and mixed-donor PBMC sciCUT&Tag experiments have been deposited as paired-end fastq files that retain the unique cellular barcodes in the Gene Expression Omnibus accession GSE224579.
Code availability
Custom code used for demultiplexing, concatenating and aligning to hg38 and mm10 reference genomes for the barnyard experiment (Snakemake41), and running Souporcell24 (Nextflow26) as well as code for generating figures is available at https://github.com/FredHutch/sciCnT_manuscript_2023. Processed data used for making figures is available at https://doi.org/10.5281/zenodo.7884460. Aggregated sciCnT PBMC coverage profiles and peak calls are interactively available at https://genome.ucsc.edu/s/jegreene/sciCnT_HOX.
References
Talbert, P. B. & Henikoff, S. The yin and yang of histone marks in transcription. Annu. Rev. Genomics Hum. Genet. 22, 147–170 (2021).
Blanco, M. A. et al. Chromatin-state barriers enforce an irreversible mammalian cell fate decision. Cell Rep. 37, 109967 (2021).
Kaya-Okur, H. S. et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10, 1930 (2019).
Bartosovic, M., Kabbe, M. & Castelo-Branco, G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 39, 825–835 (2021).
Wu, S. J. et al. Single-cell CUT&Tag analysis of chromatin modifications in differentiation and tumor progression. Nat. Biotechnol. 39, 819–824 (2021).
Zhu, C. et al. Joint profiling of histone modifications and transcriptome in single cells from mouse brain. Nat. Methods 18, 283–292 (2021).
Zhang, B. et al. Characterizing cellular heterogeneity in chromatin state with scCUT&Tag-pro. Nat. Biotechnol. 40, 1220–1230 (2022).
Kundaje, A. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Meers, M. P., Llagas, G., Janssens, D. H., Codomo, C. A. & Henikoff, S. Multifactorial profiling of epigenetic landscapes at single-cell resolution using MulTI-Tag. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01522-9 (2022).
Cao, J. et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017).
Cusanovich, D. A. et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015).
Bartlett, D. A. et al. High-throughput single-cell epigenomic profiling by targeted insertion of promoters (TIP-seq). J. Cell Biol. 220, e202103078 (2021).
Wang, Q. et al. CoBATCH for high-throughput single-cell epigenomic profiling. Mol. Cell 76, 206–216.e207 (2019).
Martin, B. K. et al. Optimized single-nucleus transcriptional profiling by combinatorial indexing. Nat. Protoc. https://doi.org/10.1038/s41596-022-00752-0 (2022).
Del Priore, I. et al. Protocol for single-cell ATAC sequencing using combinatorial indexing in mouse lung adenocarcinoma. STAR Protoc. 2, 100583 (2021).
Mezger, A. et al. High-throughput chromatin accessibility profiling at single-cell resolution. Nat. Commun. 9, 3647 (2018).
Kaya-Okur, H. S., Janssens, D. H., Henikoff, J. G., Ahmad, K. & Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264–3283 (2020).
Ren, X. et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell 184, 1895–1913 e1819 (2021).
Granja, J. M. et al. Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat. Biotechnol. 37, 1458–1465 (2019).
Chen, C. et al. Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood 139, 2198–2211 (2022).
Bartosovic, M. & Castelo-Branco, G. Multimodal chromatin profiling using nanobody-based single-cell CUT&Tag. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01535-4 (2022).
Stuart, T. et al. Nanobody-tethered transposition enables multifactorial chromatin profiling at single-cell resolution. Nat. Biotechnol. https://doi.org/10.1038/s41587-022-01588-5 (2022).
Fujiwara, Y. & Okada, Y. CUT&Tag using “stress-free” Con A-conjugated dynabeads((R)). Methods Mol. Biol. 2519, 141–153 (2023).
Heaton, H. et al. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods 17, 615–620 (2020).
Ewels, P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278 (2020).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
McInnes, L., Healy, J., & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. J. Open Source Softw. 3, 861 (2018).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Janssens, D. H. et al. Automated CUT&Tag profiling of chromatin heterogeneity in mixed-lineage leukemia. Nat. Genet. 53, 1586–1596 (2021).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Meers, M. P., Tenenbaum, D. & Henikoff, S. Peak calling by sparse enrichment analysis for CUT&RUN chromatin profiling. Epigenetics Chromatin 12, 42 (2019).
Dou, Y. et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 13, 713–719 (2006).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Lebert-Ghali, C.-É. et al. Hoxa cluster genes determine the proliferative activity of adult mouse hematopoietic stem and progenitor cells. Blood 127, 87–90 (2016).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Simon, J. A. & Kingston, R. E. Occupying chromatin: polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell 49, 808–824 (2013).
Wysocka, J. et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 442, 86–90 (2006).
Cruz, C. et al. Tri-methylation of histone H3 lysine 4 facilitates gene expression in ageing cells. eLife 7, e34081 (2018).
Mölder, F. et al. Sustainable data analysis with Snakemake. F1000 Res. https://doi.org/10.12688/f1000research.29032.2 (2021).
Acknowledgements
We thank members of our laboratory and colleagues at the Fred Hutch for providing valuable discussions and input as we developed this protocol. Specifically, we thank Fred Hutchinson Cancer Center Genomics and Bioinformatics Core Facility for sequencing expertise and services; M. Fitzgibbons for help with demultiplexing, J. Henikoff for help with genomic alignment; M. Setty, D.Otto, A.-L. Doebley and O. Waltner for bioinformatics advice; S. Bhise for help with preparing the human PBMC samples; and J. Cherone, P. Zrazhevskiy and the Andrew Stergachis laboratory for their helpful feedback on the manuscript. We are especially grateful to the many Protocols.io subscribers around the world who have tried CUT&Tag and provided helpful comments and feedback that have enriched this protocol. This work was supported by the Howard Hughes Medical Institute (S.H.), grant R01 HG010492 (S.H.) from the National Institutes of Health, and a Hartwell Foundation Postdoctoral Fellowship (D.H.J.).
Author information
Authors and Affiliations
Contributions
D.H.J., J.E.G., S.J.W., K.A. and S.H. conceived the study. J.E.G., D.H.J., S.J.W. and C.A.C. optimized the sciCUT&Tag protocol on the ICELL8. J.E.G. and D.H.J. designed the experiments and carried them out with the help of C.A.C. D.H.J. and J.E.G. performed the data analysis and generated the figures. S.N.F. provided the human PBMC samples, advised on the implementation of Souporcell for doublet detection and provided helpful discussion. S.S.M. constructed an easy-to-use Nextflow pipeline for running Souporcell. J.E.G., D.H.J., C.A.C., K.A. and S.H. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.H. has filed patent applications on related work. The other authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Julia Zeitlinger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Kaya-Okur, H. S. et al. Nat. Commun. 10, 1930 (2019): https://doi.org/10.1038/s41467-019-09982-5
Wu, S. J. et al. Nat. Biotechnol. 39, 819–824 (2021): https://doi.org/10.1038/s41587-021-00865-z
Meers, M. P. et al. Nat. Biotechnol. 41, 708–716 (2023): https://doi.org/10.1038/s41587-022-01522-9
Supplementary information
Supplementary Information
Supplementary Methods and Figs. 1 and 2.
Supplementary Table 1
All the barcoded Tn5 s5 and s7 and universal MErev adapter sequences are shown in the first tab, the barcoded i5 and i7 primer sequences in the second tab and the custom primer sequences used for illumine sequencing in the third tab.
Supplementary Table 2
Layout of the 96-well sci-pA–Tn5 plate
Supplementary Table 3
Layout of the 384-well i5 and i7 primer plate for dispensing on the ICell8 instrument
Supplementary Table 4
An example of the input file for the sciCTextract custom software that assigns sequencing reads to sample name prefixes based on the s5 and s7 adapter sequence
Supplementary Table 5
An example of the input file for the sciCTextract custom software that assigns sequencing reads to sample name suffixes based on the i5 and i7 adapter sequence
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Janssens, D.H., Greene, J.E., Wu, S.J. et al. Scalable single-cell profiling of chromatin modifications with sciCUT&Tag. Nat Protoc 19, 83–112 (2024). https://doi.org/10.1038/s41596-023-00905-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00905-9
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.