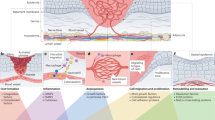
Abstract
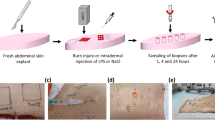
Wound healing is a complex physiological process involving various cell types and signaling pathways. The capability to observe the dynamics of wound repair offers valuable insights into the effects of genetic modifications, pharmaceutical interventions or other experimental manipulations on the skin-repair process. Here, we provide a comprehensive protocol for a full-thickness, excisional skin-wound-healing assay in mice, which can easily be performed by any scientist who has received an animal welfare course certificate and can be completed within ~3 h, depending on the number of animals. Crucially, we highlight the importance of considering key aspects of the assay that can dramatically contribute to the reliability and reproducibility of these experiments. We thoroughly discuss the experimental design, necessary preparations, wounding technique and analysis. In addition, we discuss the use of lineage-tracing techniques to monitor cell migration, differentiation and the contribution of different cell populations to the repair process. Overall, we explore key aspects of the skin-wound-healing assay, supplying a detailed procedure and guidelines essential for decreasing variability and obtaining reliable and reproducible results.
Key points
-
The protocol provides an accurate method for performing a full-thickness excisional wound-healing (WH) assay in murine dorsal skin with high reproducibility.
-
This method can improve WH studies by decreasing deviations and improving reliability, eventually reducing the number of animals required per experiment. In addition, the assay can be combined with lineage-tracing experiments to investigate the contribution of different cell populations to the repair process.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All raw data, including measurements for Supplementary Fig. 1b and original images for Fig. 4, are available in Supplementary Data 2. Source data are provided with this paper.
References
Koren, E. et al. Thy1 marks a distinct population of slow-cycling stem cells in the mouse epidermis. Nat. Commun. 13, 4628 (2022).
Aragona, M. et al. Defining stem cell dynamics and migration during wound healing in mouse skin epidermis. Nat. Commun. 8, 14684 (2017).
Lisse, T. S., Sharma, M., Vishlaghi, N., Pullagura, S. R. & Braun, R. E. GDNF promotes hair formation and cutaneous wound healing by targeting bulge stem cells. NPJ Regen. Med. 5, 13 (2020).
Dekoninck, S. & Blanpain, C. Stem cell dynamics, migration and plasticity during wound healing. Nat. Cell Biol. 21, 18–24 (2019).
Chou, W. C. et al. Direct migration of follicular melanocyte stem cells to the epidermis after wounding or UVB irradiation is dependent on Mc1r signaling. Nat. Med. 19, 924–929 (2013).
Page, M. E., Lombard, P., Ng, F., Göttgens, B. & Jensen, K. B. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell 13, 471–482 (2013).
Donati, G. et al. Wounding induces dedifferentiation of epidermal Gata6+ cells and acquisition of stem cell properties. Nat. Cell Biol. 19, 603–613 (2017).
Lu, C. P. et al. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 150, 136–150 (2012).
Shook, B. A. et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 362, eaar2971 (2018).
Plikus, M. V. et al. Regeneration of fat cells from myofibroblasts during wound healing. Science 355, 748–752 (2017).
Shook, B. A. et al. Dermal adipocyte lipolysis and myofibroblast conversion are required for efficient skin repair. Cell Stem Cell 26, 880–895.e6 (2020).
McGee, H. M. et al. IL-22 promotes fibroblast-mediated wound repair in the skin. J. Invest. Dermatol. 133, 1321–1329 (2013).
Shook, B., Xiao, E., Kumamoto, Y., Iwasaki, A. & Horsley, V. CD301b+ macrophages are essential for effective skin wound healing. J. Invest. Dermatol. 136, 1885–1891 (2016).
Toulon, A. et al. A role for human skin-resident T cells in wound healing. J. Exp. Med. 206, 743–750 (2009).
Keyes, B. E. et al. Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell 167, 1323–1338.e14 (2016).
Lim, C. H. et al. Hedgehog stimulates hair follicle neogenesis by creating inductive dermis during murine skin wound healing. Nat. Commun. 9, 4903 (2018).
Wietecha, M. S. et al. Activin-mediated alterations of the fibroblast transcriptome and matrisome control the biomechanical properties of skin wounds. Nat. Commun. 11, 2604 (2020).
Rognoni, E. et al. Inhibition of β-catenin signalling in dermal fibroblasts enhances hair follicle regeneration during wound healing. Development 143, 2522–2535 (2016).
Hiebert, P. et al. Nrf2-mediated fibroblast reprogramming drives cellular senescence by targeting the matrisome. Dev. Cell 46, 145–161.e10 (2018).
Naik, S. et al. Inflammatory memory sensitizes skin epithelial stem cells to tissue damage. Nature 550, 475–480 (2017).
Gonzales, K. A. U. et al. Stem cells expand potency and alter tissue fitness by accumulating diverse epigenetic memories. Science 374, eabh2444 (2021).
Chen, X. et al. IL-17R–EGFR axis links wound healing to tumorigenesis in Lrig1+ stem cells. J. Exp. Med. 216, 195–214 (2019).
Weber, C. et al. Macrophage infiltration and alternative activation during wound healing promote MEK1-induced skin carcinogenesis. Cancer Res. 76, 805–817 (2016).
Ge, Y. et al. Stem cell lineage infidelity drives wound repair and cancer. Cell 169, 636–650.e14 (2017).
Lintel, H. et al. Transdermal deferoxamine administration improves excisional wound healing in chronically irradiated murine skin. J. Transl. Med. 4, 1–13 (2022).
Ben-Yehuda Greenwald, M. et al. Topical wound treatment with a nitric oxide-releasing PDE5 inhibitor formulation enhances blood perfusion and promotes healing in mice. Pharmaceutics 14, 2358 (2022).
Ankawa, R. et al. Apoptotic cells represent a dynamic stem cell niche governing proliferation and tissue regeneration. Dev. Cell 56, 1900–1916.e5 (2021).
Fuchs, Y. et al. Sept4/ARTS regulates stem cell apoptosis and skin regeneration. Science 341, 286–289 (2013).
Sedov, E. et al. THY1-mediated mechanisms converge to drive YAP activation in skin homeostasis and repair. Nat. Cell Biol. 24, 1049–1063 (2022).
Rossiter, H. et al. Loss of vascular endothelial growth factor A activity in murine epidermal keratinocytes delays wound healing and inhibits tumor formation. Cancer Res. 64, 3508–3516 (2004).
Nguyen, H. et al. Tcf3 and Tcf4 are essential for long-term homeostasis of skin epithelia. Nat. Genet. 41, 1068–1075 (2009).
Jirkof, P., Cesarovic, N., Rettich, A., Fleischmann, T. & Arras, M. Individual housing of female mice: influence on postsurgical behaviour and recovery. Lab. Anim. 46, 325–334 (2012).
Pastar, I. et al. Epithelialization in wound healing: a comprehensive review. Adv. Wound Care 3, 445–464 (2014).
Foster, D. S. et al. Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proc. Natl Acad. Sci. USA 118, e2110025118 (2021).
Talbott, H. E., Mascharak, S., Griffin, M., Wan, D. C. & Longaker, M. T. Wound healing, fibroblast heterogeneity, and fibrosis. Cell Stem Cell 29, 1161–1180 (2022).
Chen, L., Mirza, R., Kwon, Y., DiPietro, L. A. & Koh, T. J. The murine excisional wound model: contraction revisited. Wound Repair Regen. 23, 874–877 (2015).
Hinz, B. Formation and function of the myofibroblast during tissue repair. J. Invest. Dermatol. 127, 526–537 (2007).
Darby, I. A., Laverdet, B., Bonté, F. & Desmoulière, A. Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 7, 301–311 (2014).
Mascharak, S. et al. Multi-omic analysis reveals divergent molecular events in scarring and regenerative wound healing. Cell Stem Cell 29, 315–327.e6 (2022).
Kurita, M. et al. In vivo reprogramming of wound-resident cells generates skin epithelial tissue. Nature 561, 243–247 (2018).
Mascharak, S. et al. Preventing Engrailed-1 activation in fibroblasts yields wound regeneration without scarring. Science 372, eaba2374 (2021).
Lumbers, M. Understanding and addressing dryness during wound healing. Br. J. Community Nurs. 24, S11–S14 (2019).
Davidson, J. M., Yu, F. & Opalenik, S. R. Splinting strategies to overcome confounding wound contraction in experimental animal models. Adv. Wound Care 2, 142–148 (2013).
Wang, X., Ge, J., Tredget, E. E. & Wu, Y. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc. 8, 302–309 (2013).
Ganguli-Indra, G. Protocol for cutaneous wound healing assay in a murine model. in Stem Cells and Tissue Repair: Methods and Protocols (ed. Kioussi, C.) 151–159 (Humana Press, 2014).
Falanga, V. et al. Full-thickness wounding of the mouse tail as a model for delayed wound healing: accelerated wound closure in Smad3 knock-out mice. Wound Repair Regen. 12, 320–326 (2004).
Gurtner, G. C., Wong, V. W., Sorkin, M., Glotzbach, J. P. & Longaker, M. T. Surgical approaches to create murine models of human wound healing. J. Biomed. Biotechnol. 2011, 969618 (2011).
Landén, N. X., Li, D. & Ståhle, M. Transition from inflammation to proliferation: a critical step during wound healing. Cell. Mol. Life Sci. 73, 3861–3885 (2016).
Galiano, R. D., Michaels, J. V, Dobryansky, M., Levine, J. P. & Gurtner, G. C. Quantitative and reproducible murine model of excisional wound healing. Wound Repair Regen. 12, 485–492 (2004).
Dabiri, G., Damstetter, E. & Phillips, T. Choosing a wound dressing based on common wound characteristics. Adv. Wound Care 5, 32–41 (2016).
Jones, V., Grey, J. E. & Harding, K. G. Wound dressings. BMJ 332, 777–780 (2006).
Baranoski, S. & Ayello, E. A. Wound dressings: an evolving art and science. Adv. Skin. Wound Care 25, 87–92 (2012).
Winter, G. D. Formation of the scab and the rate of epithelization of superficial wounds in the skin of the young domestic pig. Nature 193, 293–294 (1962).
Charan, J. & Kantharia, N. How to calculate sample size in animal studies? J. Pharmacol. Pharmacother. 4, 303–306 (2013).
Festing, M. F. W. & Altman, D. G. Guidelines for the design and statistical analysis of experiments using laboratory animals. ILAR J. 43, 244–257 (2002).
Jaks, V. et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40, 1291–1299 (2008).
Ansell, D. M., Kloepper, J. E., Thomason, H. A., Paus, R. & Hardman, M. J. Exploring the “hair growth–wound healing connection”: anagen phase promotes wound re-epithelialization. J. Invest. Dermatol. 131, 518–528 (2011).
Müller-Röver, S. et al. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 117, 3–15 (2001).
Potten, C. S. Some observations on the post-plucking depression in tritiated thymidine utilization in mouse skin and some tentative cell kinetic determinations. J. Invest. Dermatol. 58, 180–185 (1972).
Chen, C. C. et al. Organ-level quorum sensing directs regeneration in hair stem cell populations. Cell 161, 277–290 (2015).
Silver, A.F. & Chase, H. B. Early anagen initiated by plucking compared with early spontaneous anagen. In Advances in Biology of Skin. Vol. 9 265–286 (Pergamon Press, 1967).
Stojadinovic, O., Ito, M. & Tomic-Canic, M. Hair cycling and wound healing: to pluck or not to pluck? J. Invest. Dermatol. 131, 292–294 (2011).
Zhang, B. et al. Hyperactivation of sympathetic nerves drives depletion of melanocyte stem cells. Nature 577, 676–681 (2020).
Guarnieri, M. et al. Safety and efficacy of buprenorphine for analgesia in laboratory mice and rats. Lab Anim. (NY). 41, 337–343 (2012).
Bravo, L., Mico, J. A. & Berrocoso, E. Discovery and development of tramadol for the treatment of pain. Expert Opin. Drug Discov. 12, 1281–1291 (2017).
Clark, T. S., Clark, D. D. & Hoyt, R. F. Pharmacokinetic comparison of sustained-release and standard buprenorphine in mice. J. Am. Assoc. Lab. Anim. Sci. 53, 387–391 (2014).
Van Loo, P. L., Mol, J. A., Koolhaas, J. M., Van Zutphen, B. F. & Baumans, V. Modulation of aggression in male mice: influence of group size and cage size. Physiol. Behav. 72, 675–683 (2001).
Glasper, E. R. & DeVries, A. C. Social structure influences effects of pair-housing on wound healing. Brain Behav. Immun. 19, 61–68 (2005).
Pyter, L. M. et al. Contrasting mechanisms by which social isolation and restraint impair healing in male mice. Stress 17, 256–265 (2014).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Nagy, A. Cre recombinase: the universal reagent for genome tailoring. Genesis 26, 99–109 (2000).
Vasioukhin, V., Degenstein, L., Wise, B. & Fuchs, E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl Acad. Sci. USA 96, 8551–8556 (1999).
Jahn, H. M. et al. Refined protocols of tamoxifen injection for inducible DNA recombination in mouse astroglia. Sci. Rep. 8, 5913 (2018).
Horng, H. C. et al. Estrogen effects on wound healing. Int. J. Mol. Sci. 18, 1–14 (2017).
Irrera, N. et al. Dietary management of skin health: the role of genistein. Nutrients 9, 1–10 (2017).
Kellendonk, C. et al. Regulation of Cre recombinase activity by the synthetic steroid RU 486. Nucleic Acids Res. 24, 1404–1411 (1996).
Berton, T. R. et al. Characterization of an inducible, epidermal-specific knockout system: differential expression of lacZ in different Cre reporter mouse strains. Genesis 26, 160–161 (2000).
Morris, R. J. et al. Capturing and profiling adult hair follicle stem cells. Nat. Biotechnol. 22, 411–417 (2004).
He, F. & Soriano, P. Sox10ERT2CreERT2 mice enable tracing of distinct neural crest cell populations. Dev. Dyn. 244, 1394–1403 (2015).
Wang, X. et al. Macrophages induce AKT/β-catenin-dependent Lgr5+ stem cell activation and hair follicle regeneration through TNF. Nat. Commun. 8, 14091 (2017).
Ruzankina, Y. et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 1, 113–126 (2007).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 1–8 (2001).
Arras, M., Autenried, P., Rettich, A., Spaeni, D. & Rülicke, T. Optimization of intraperitoneal injection anesthesia in mice: drugs, dosages, adverse effects, and anesthesia depth. Comp. Med. 51, 443–456 (2001).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Ankawa, R. & Fuchs, Y. May the best wound WIHN: the hallmarks of wound-induced hair neogenesis. Curr. Opin. Genet. Dev. 72, 53–60 (2022).
Driskell, R. R. et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281 (2013).
Acknowledgements
We thank current and former members of the Fuchs group for their insights and experience. We thank M. Yosopuva, R. Ankawa and N. Goldberger, for their technical assistance; V. Zlobin and Y. Nissan for animal house support; and N. Dahan and Y. Lupu-haber for insights regarding microscopy. We extend special thanks to D. Kulinsky and A. Hezi-Yamit for valuable input. Y.F. is supported by the EMBO Young Investigator program.
Author information
Authors and Affiliations
Contributions
M.Y. and Y.F. designed, analyzed and conceived the project. M.Y. performed imaging. M.Y, I.B. and Y.F. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
M.Y., I.B. and Y.F. are employees and/or shareholders of Augmanity, a privately funded research institute in Rehovot, Israel. The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Koren, E. et al. Nat. Commun. 13, 4628 (2022): https://doi.org/10.1038/s41467-022-31629-1
Ankawa, R. et al. Dev. Cell 56, 1900–1916.e5 (2021): https://doi.org/10.1016/j.devcel.2021.06.008
Extended data
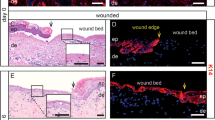
Extended Data Fig. 1 Whole-mount preparation of a sealed wound.
a, HFs in the vicinity of the wound edge enter the anagen phase, extending to the hypodermis and hindering epidermis-dermis separation. b, Scar tissue (30 d after wound inflection) is scrapped from fat and immersed entirely in EDTA to weaken the epidermal-dermal connection. c, Under a stereomicroscope, using two pairs of forceps, the epidermis is gently separated from the dermis. d, The dermis is initially fixed with a low volume of fixative to prevent the tissue from folding.
Extended Data Fig. 2 Calculating the wound area by using the Fiji (Image J) Software.
a–c, Open high-resolution scan in Fiji (a) and select the ‘Make Binary’ command to transform the image to white & black (b and c). The red square in c defines the selected zoomed-in area in D. d–g, Zoom-in to detect small gaps in the outlines (the purple-dashed square marks the gapped area). Use the hand-free selection tool (d, blue square) and the ‘Clear’ command to fill the gaps. Use the ‘Wand’ tool (g, blue square) to select the area you wish to calculate and press ‘M’ on your keyboard to perform the measurements. The values will be shown in a pop-up window and can be copied directly to an Excel file.
Supplementary information
Supplementary Information
Supplementary Protocols 1–3, Discussion and Figs. 1 and 2
Supplementary Data 1
Theoretical data analysis template
Supplementary Data 2
Source data for Supplementary Fig. 1B (graph)
Source data
Source Data Fig. 4
Original images
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yampolsky, M., Bachelet, I. & Fuchs, Y. Reproducible strategy for excisional skin-wound-healing studies in mice. Nat Protoc 19, 184–206 (2024). https://doi.org/10.1038/s41596-023-00899-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00899-4
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.