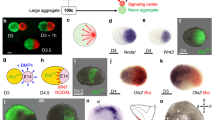
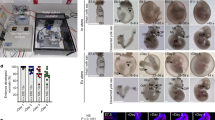
Abstract
The interaction between embryonic and extraembryonic tissues is critical in natural mouse embryogenesis. Here, to enable such interaction in vitro, we describe a protocol to assemble a complete mouse embryo model using mouse embryonic stem cells and induced embryonic stem cells to express Cdx2 (or trophoblast stem cells) and Gata4 to reconstitute the epiblast, extraembryonic ectoderm and visceral endoderm lineages, respectively. The resulting complete embryo models recapitulate development from embryonic day 5.0 to 8.5, generating advanced embryonic and extraembryonic tissues that develop through gastrulation to initiate organogenesis to form a head and a beating heart structure as well as a yolk sac and chorion. Once the required stem cell lines are stably maintained in culture, the protocol requires 1 day to assemble complete embryo models and a further 8 days to culture them until headfold stages, although structures can be collected at earlier developmental stages as required. This protocol can be easily performed by researchers with experience in mouse stem cell culture, although they will benefit from knowledge of natural mouse embryos at early postimplantation stages.
Key points
-
This protocol is for the assembly of complete mouse embryo models from embryonic and induced stem cells. These embryo models develop to generate advanced embryonic and extraembryonic tissues.
-
While there are many existing in vitro models of mouse embryogenesis, these complete induced embryo models are among the only models reported to recapitulate development from gastrulation to headfold stages.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009).
Xu, P. F. et al. Construction of a mammalian embryo model from stem cells organized by a morphogen signalling centre. Nat. Commun. 12, 1–22 (2021).
van den Brink, S. C. et al. Single-cell and spatial transcriptomics reveal somitogenesis in gastruloids. Nature 582, 405–409 (2020).
Rossi, G. et al. Capturing cardiogenesis in gastruloids. Cell Stem Cell 28, 230–240.e6 (2020).
Beccari, L. et al. Multi-axial self-organization properties of mouse embryonic stem cells into gastruloids. Nature 562, 272–276 (2018).
Veenvliet, J. V. et al. Mouse embryonic stem cells self-organize into trunk-like structures with neural tube and somites. Science 370, eaba4937 (2020).
Amadei, G. et al. Embryo model completes gastrulation to neurulation and organogenesis. Nature 610, 143–153 (2022).
Lau, K. Y. C. et al. Mouse embryo model derived exclusively from embryonic stem cells undergoes neurulation and heart development. Cell Stem Cell 29, 1445–1458.e8 (2022).
Harrison, S. E., Sozen, B., Christodoulou, N., Kyprianou, C. & Zernicka-Goetz, M. Assembly of embryonic and extraembryonic stem cells to mimic embryogenesis in vitro. Science 356, eaal1810 (2017).
Harrison, S. E., Sozen, B. & Zernicka-Goetz, M. In vitro generation of mouse polarized embryo-like structures from embryonic and trophoblast stem cells. Nat. Protoc. 13, 1586–1602 (2018).
Sozen, B. et al. Self-assembly of embryonic and two extra-embryonic stem cell types into gastrulating embryo-like structures. Nat. Cell Biol. 20, 979–989 (2018).
Shimosato, D., Shiki, M. & Niwa, H. Extra-embryonic endoderm cells derived from ES cells induced by GATA Factors acquire the character of XEN cells. BMC Dev. Biol. 7, 80 (2007).
Amadei, G. et al. Inducible stem-cell-derived embryos capture mouse morphogenetic events in vitro. Dev. Cell 56, 366–382.e9 (2021).
Sturm, K. & Tam, P. P. L. Isolation and culture of whole postimplantation embryos and germ layer derivatives. Methods Enzymol. 225, 164–190 (1993).
Aguilera-Castrejon, A. et al. Ex utero mouse embryogenesis from pre-gastrulation to late organogenesis. Nature 593, 119–124 (2021).
Chawengsaksophak, K., James, R., Hammond, V. E., Köntgen, F. & Beck, F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 386, 84–87 (1997).
Strumpf, D. et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 132, 2093–2102 (2005).
Jedrusik, A. et al. Role of Cdx2 and cell polarity in cell allocation and specification of trophectoderm and inner cell mass in the mouse embryo. Genes Dev. 22, 2692–2706 (2008).
Niwa, H. et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell 123, 917–929 (2005).
Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. Promotion to trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 (1998).
Quinn, J., Kunath, T. & Rossant, J. Mouse trophoblast stem cells. Methods Mol. Med. 121, 125–148 (2006).
Seong, J. et al. Epiblast inducers capture mouse trophectoderm stem cells in vitro and pattern blastoids for implantation in utero. Cell Stem Cell 29, 1102–1118.e8 (2022).
Tarazi, S. et al. Post-gastrulation synthetic embryos generated ex utero from mouse naive ESCs. Cell 185, 3290–3306.e25 (2022).
Acknowledgements
The authors thank S. Raj for providing the images for Fig. 3, K. Ivanovitch for advice on the handling and storage of rat serum and M.Z.-G. laboratory members for their helpful comments. This work was supported by National Institutes of Health Pioneer Award (DP1 HD104575-01), the Allen Discovery Center for Lineage Tracing; European Research Council (669198), the Wellcome Trust (207415/Z/17/Z), Open Philanthropy/Silicon Valley Community Foundation and Weston Havens Foundation. K.Y.C.L. was supported by the Croucher Foundation and the Cambridge Trust.
Author information
Authors and Affiliations
Contributions
K.Y.C.L. performed the experiments. K.Y.C.L. and G.A. developed the protocol with the guidance from M.Z.-G. M.Z.-G. conceived and supervised the project. K.Y.C.L., G.A. and M.Z.-G. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
A patent ‘Generation of synthetic embryos from multiple stem cell types’ was filed by California Institute of Technology and the University of Cambridge under CIT file number: CIT-8826-P and serial number: 63/344,251.
Peer review
Peer review information
Nature Protocols thanks Miki Ebisuya and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Amadei, G. et al. Nature 610, 143–153 (2022): https://doi.org/10.1038/s41586-022-05246-3
Amadei, G. et al. Dev. Cell 56, 366–382.e9 (2021): https://doi.org/10.1016/j.devcel.2020.12.004
Lau, K. Y. C. et al. Cell Stem Cell 29, 1445–1458.e8 (2022): https://doi.org/10.1016/j.stem.2022.08.013
Rights and permissions
About this article
Cite this article
Lau, K.Y.C., Amadei, G. & Zernicka-Goetz, M. Assembly of complete mouse embryo models from embryonic and induced stem cell types in vitro. Nat Protoc 18, 3662–3689 (2023). https://doi.org/10.1038/s41596-023-00891-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00891-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.