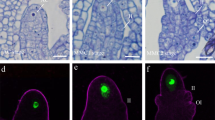
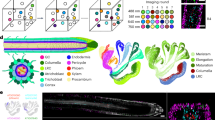
Abstract
RNA-sequencing (RNA-seq) provides invaluable knowledge on developmental pathways and the effects of mutant phenotypes. Plant reproductive cells have traditionally been difficult to isolate for genomics because they are rare and often deeply embedded within somatic tissues. Here, we present a protocol to isolate single maize meiocytes and pollen grains for RNA-seq. We discuss how to identify and isolate each sample type under a microscope, prepare RNA-seq libraries and analyze the data. This technique has several advantages over alternative methods, combining the ability to target specific rare cell types while resolving cell-to-cell heterogeneity with single-cell RNA-seq. The technique is compatible with minute amounts of starting material (e.g., a single anther), making it possible to collect dense time courses. Furthermore, developmentally synchronized anthers are saved for microscopy, allowing staging to be performed in parallel with expression analysis. Up to 200 cells can be collected in 4–5 h by someone proficient in tissue dissection, and library preparation can be completed in 2 d by researchers experienced in molecular biology and genomics. This protocol will facilitate research on plant reproduction, providing insights critical to plant breeding, genetics and agriculture.
Key points
-
In this protocol, maize meiocytes and pollen grains are microscopically identified and isolated for single-cell RNA-seq by using a modified CEL-Seq2 workflow. Developmental staging of anthers is also performed in parallel with expression analysis.
-
Rare reproductive cells are specifically targeted, and larger cell sizes and sample numbers are accommodated than in droplet-based single-cell RNA-seq methods. This protocol is compatible with minute amounts of material and resolves heterogeneity by focusing on single cells and pollen grains.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The example sequencing data analyzed in this protocol are available on the Gene Expression Omnibus under accession GSE121039. The data in Fig. 5 were reanalyzed from refs. 3,4 (Gene Expression Omnibus accessions GSE121039 and GSE175505). The ‘data analysis’ section can be followed by using example data from ref. 3, which can be accessed on the Sequence Read Archive under accessions SRR7989735 and SRR7989736.
References
Ozias-Akins, P. & Conner, J. A. Clonal reproduction through seeds in sight for crops. Trends Genet. 36, 215–226 (2020).
Muñoz-Sanz, J. V., Zuriaga, E., Cruz-García, F., McClure, B. & Romero, C. Self-(in)compatibility systems: target traits for crop production, plant breeding, and biotechnology. Front. Plant Sci. 11, 195 (2020).
Nelms, B. & Walbot, V. Defining the developmental program leading to meiosis in maize. Science 364, 52–56 (2019).
Nelms, B. & Walbot, V. Gametophyte genome activation occurs at pollen mitosis I in maize. Science 375, 424–429 (2022).
Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 (2016).
Kivioja, T. et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2011).
MacConaill, L. E. et al. Unique, dual-indexed sequencing adapters with UMIs effectively eliminate index cross-talk and significantly improve sensitivity of massively parallel sequencing. BMC Genomics 19, 30 (2018).
Sinha, R. et al. Index switching causes ‘spreading-of-signal’ among multiplexed samples in Illumina HiSeq 4000 DNA sequencing. Preprint at https://www.biorxiv.org/content/10.1101/125724v1 (2017).
Bennett, M. D. The duration of meiosis. Proc. R. Soc. Lond. B 178, 277–299 (1971).
Nonomura, K. I. et al. A novel RNA-recognition-motif protein is required for premeiotic G1/S-phase transition in rice (Oryza sativa L.). PLoS Genet. 7, e1001265 (2011).
Li, J. et al. The plant-specific protein FEHLSTART controls male meiotic entry, initializing meiotic synchronization in Arabidopsis. Plant J. 84, 659–671 (2015).
Li, X., Li, L. & Yan, J. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6, 6648 (2015).
Xu, W. et al. A plate-based single-cell ATAC-seq workflow for fast and robust profiling of chromatin accessibility. Nat. Protoc. 16, 4084–4107 (2021).
Zheng, G. X. Y. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Englhart, M., Šoljić, L. & Sprunck, S. Manual isolation of living cells from the Arabidopsis thaliana female gametophyte by micromanipulation. Methods Mol. Biol. 1669, 221–234 (2017).
Dukowic-Schulze, S. et al. The transcriptome landscape of early maize meiosis. BMC Plant Biol. 14, 118 (2014).
Borg, M. et al. Epigenetic reprogramming rewires transcription during the alternation of generations in Arabidopsis. Elife 10, e61894 (2021).
Tang, X. et al. Global gene profiling of laser-captured pollen mother cells indicates molecular pathways and gene subfamilies involved in rice meiosis. Plant Physiol. 154, 1855–1870 (2010).
Zhang, H. et al. Transcriptomes and proteomes define gene expression progression in pre-meiotic maize anthers. G3 (Bethesda) 4, 993–1010 (2014).
Ramsköld, D. et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782 (2012).
Hagemann-Jensen, M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020).
Srivastava, A., Malik, L., Smith, T., Sudbery, I. & Patro, R. Alevin efficiently estimates accurate gene abundances from dscRNA-seq data. Genome Biol. 20, 65 (2019).
Dawe, R. K., Sedat, J. W., Agard, D. A. & Cande, W. Z. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76, 901–912 (1994).
Illumina Knowledge Article #1252: best practices for manually normalizing library concentrations. Illumina, Inc. (21 September 2023); https://knowledge.illumina.com/library-preparation/general/library-preparation-general-reference_material-list/000001252.
Steffen, J. G., Kang, I.-H., Macfarlane, J. & Drews, G. N. Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292 (2007).
Acknowledgements
The authors thank V. Walbot, who provided invaluable guidance during the development of this technique, and J. Gent for his helpful feedback and suggestions. This work was supported by NSF grant no. 2218712 to B.N. J.A.-F. was supported by CONACyT PhD fellowship 781792 and by CONACyT Ciencia Básica 2017-2018 grant A1-S-34956.
Author information
Authors and Affiliations
Contributions
All authors contributed to the development and writing of this protocol.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Chloé Girard, Marc Libault and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol:
Nelms, B. & Walbot, V. Science 364, 52–56 (2019): https://doi.org/10.1126/science.aav6428
Nelms, B. & Walbot, V. Science 375, 424–429 (2022): https://doi.org/10.1126/science.abl7392
Supplementary information
Supplementary Information
Supplementary Figs. 1 and 2 and Supplementary Table 2
Supplementary Table
Supplementary Table 1. Sequence of barcoded CEL-seq primers. The first of these primers is shown in Supplementary Figure 2a with each section annotated. This table can be directly uploaded to IDT for ordering plate-based oligos. Because there are randomized bases in these primers (Ns), the automated form will ask if you want ‘machine-mixed’ or ‘hand-mixed’ bases during ordering; select machine-mixed bases.
Supplementary Data 1
Supplementary Data 1. CELseq_whitelist_legacy.txt. Legacy CELseq whitelist for Salmon Alevin. Use when re-analyzing data from ref. 3.
Supplementary Data 2
Supplementary Data 2. TGMAP.tsv. Transcript to gene mapping for Salmon Alevin.
Supplementary Data 3
Supplementary Data 3. CELseq_whitelist.txt. CELseq whitelist for Salmon Alevin. Use when analyzing new data generated with this protocol.
Supplementary Video 1
Supplementary Video 1. Technique to press anthers and release meiotic cells or pollen precursors (Step 5A(v–vii) and 5B(ii)).
Supplementary Video 2
Supplementary Video 2. Technique to isolate single cells and pollen precursors by using a syringe needle (Steps 6–12).
Supplementary Video 3
Supplementary Video 3. Handling of single-cell material on PCR caps (Steps 14 and 22–25).
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Washburn, M., Alaniz-Fabián, J., Scroggs, T. et al. Single-cell RNA-seq of maize meiocytes and pollen grains. Nat Protoc 18, 3512–3533 (2023). https://doi.org/10.1038/s41596-023-00889-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00889-6
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.