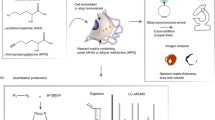
Abstract
Connective tissues are essential building blocks for organ development, repair and regeneration. However, we are at the early stages of understanding connective tissue dynamics. Here, we detail a method that enables in vivo fate mapping of organ extracellular matrix (ECM) by taking advantage of a crosslinking chemical reaction between amine groups and N-hydroxysuccinimide esters. This methodology enables robust labeling of ECM proteins, which complement previous affinity-based single-protein methods. This protocol is intended for entry-level scientists and the labeling step takes between 5 and 10 min. ECM ‘tagging’ with fluorophores using N-hydroxysuccinimide esters enables visualization of ECM spatial modifications and is particularly useful to study connective tissue dynamics in organ fibrosis, tumor stroma formation, wound healing and regeneration. This in vivo chemical fate mapping methodology is highly versatile, regardless of the tissue/organ system, and complements cellular fate-mapping techniques. Furthermore, as the basic chemistry of proteins is highly conserved between species, this method is also suitable for cross-species comparative studies of ECM dynamics.
Key points
-
This protocol describes a method for broad-spectrum, in vivo fluorescent labeling and tracking of extracellular matrix (ECM) proteins through the systemic or local application of N-hydroxysuccinimide esters. This flexible approach can be adapted to a variety of organ systems and wounding models.
-
In contrast to common ECM-tracing methods that enable labeling single ECM proteins, this N-hydroxysuccinimide ester-based chemical technique tags virtually all ECM proteins to unveil more general ECM mechanics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Baron, C. S. & van Oudenaarden, A. Unravelling cellular relationships during development and regeneration using genetic lineage tracing. Nat. Rev. Mol. Cell Biol. 20, 753–765 (2019).
Hastings, J. F., Skhinas, J. N., Fey, D., Croucher, D. R. & Cox, T. R. The extracellular matrix as a key regulator of intracellular signalling networks. Br. J. Pharmacol. 176, 82–92 (2019).
Manou, D. et al. The complex interplay between extracellular matrix and cells in tissues. Methods Mol. Biol. 1952, 1–20 (2019).
Loganathan, R. et al. Extracellular matrix motion and early morphogenesis. Development 143, 2056–2065 (2016).
Sivakumar, P. et al. New insights into extracellular matrix assembly and reorganization from dynamic imaging of extracellular matrix proteins in living osteoblasts. J. Cell Sci. 119, 1350–1360 (2006).
Aufschnaiter, R. et al. In vivo imaging of basement membrane movement: ECM patterning shapes Hydra polyps. J. Cell Sci. 124, 4027–4038 (2011).
Zamir, E. A., Czirók, A., Cui, C., Little, C. D. & Rongish, B. J. Mesodermal cell displacements during avian gastrulation are due to both individual cell-autonomous and convective tissue movements. Proc. Natl Acad. Sci. USA 103, 19806–19811 (2006).
Czirók, A., Rongish, B. J. & Little, C. D. Extracellular matrix dynamics during vertebrate axis formation. Dev. Biol. 268, 111–122 (2004).
Dzamba, B. J. & DeSimone, D. W. Extracellular matrix (ECM) and the sculpting of embryonic tissues. Curr. Top. Dev. Biol. 130, 245–274 (2018).
Duffield, M. Dynamic views. In Practical App Development with Aurelia Ch. 18, 185–194 (Apress, 2018).
Pankov, R. & Yamada, K. M. Non‐radioactive quantification of fibronectin matrix assembly. Curr. Protoc. Cell Biol. 25, 1–9 (2004).
Aper, S. J. A. et al. Colorful protein-based fluorescent probes for collagen imaging. PLoS ONE 9, 1–21 (2014).
Leonard, A. K. et al. Methods for the visualization and analysis of extracellular matrix protein structure and degradation. Methods Cell Biol. 143, 79–95 (2019).
Arnoldini, S. et al. Novel peptide probes to assess the tensional state of fibronectin fibers in cancer. Nat. Commun. 8, 1793 (2017).
Keeley, D. P. et al. Comprehensive endogenous tagging of basement membrane components reveals dynamic movement within the matrix scaffolding. Dev. Cell 54, 60–74 (2021).
Morris, J. L. et al. Live imaging of collagen deposition during skin development and repair in a collagen I–GFP fusion transgenic zebrafish line. Dev. Biol. 441, 4–11 (2018).
Ohashi, T., Kiehart, D. P. & Erickson, H. P. Dual labeling of the fibronectin matrix and actin cytoskeleton with green fluorescent protein variants. J. Cell Sci. 115, 1221–1229 (2002).
Lu, Y. et al. Live imaging of type I collagen assembly dynamics in osteoblasts stably expressing GFP and mCherry-tagged collagen constructs. J. Bone Miner. Res. 33, 1166–1182 (2018).
Poole, J. J. A. & Mostaço-guidolin, L. B. Optical microscopy and the extracellular matrix structure: a review. Cells 10, 1760 (2021).
Berg, E. A. & Fishman, J. B. Labeling antibodies. Cold Spring Harb. Protoc. 2020, 252–263 (2020).
Fischer, A. et al. Neutrophils direct preexisting matrix to initiate repair in damaged tissues. Nat. Immunol. 23, 518–531 (2022).
Correa-Gallegos, D. et al. Patch repair of deep wounds by mobilized fascia. Nature 576, 287–292 (2019).
Wan, L. et al. Connexin43 gap junction drives fascia mobilization and repair of deep skin wounds. Matrix Biol. 97, 58–71 (2021).
Kashimoto, R. et al. Lattice-patterned collagen fibers and their dynamics in axolotl skin regeneration. iScience 25, 104524 (2022).
Acknowledgements
We thank S. Sabrautzki for her veterinary advice and animal welfare support. We thank S. Dietzel and the LMU microscopy unit for their support with multi-photon imaging. Y.R. is supported by the European Research Council Consolidator Grant (ERC-CoG 819933), the LEO Foundation (LF-OC-21-000835) and the European Foundation for the Study of Diabetes (EFSD) Anniversary Fund Program.
Author information
Authors and Affiliations
Contributions
D.C.-G. developed the skin ECM labeling method. A.F. and J.W. developed the internal organ ECM labeling methods. J.W. provided veterinary advice and animal experimental protocols. S.C. assisted with multiphoton imaging. A.F., D.C.-G., H.-G.M. and Y.R. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare the following competing interests: Y.R. and A.F. have filed patent application EP21206 688.0 covering the use of these methods to study ECM movement in organ fibrosis.
Peer review
Peer review information
Nature Protocols thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Correa-Gallegos, D. et al. Nature 576, 287–292 (2019): https://doi.org/10.1038/s41586-019-1794-y
Fischer, A. et al. Nat. Immunol. 23, 518–531 (2022): https://doi.org/10.1038/s41590-022-01166-6
Wan, L. et al. Matrix Biol. 97, 58–71 (2021): https://doi.org/10.1016/j.matbio.2021.01.005
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fischer, A., Correa-Gallegos, D., Wannemacher, J. et al. In vivo fluorescent labeling and tracking of extracellular matrix. Nat Protoc 18, 2876–2890 (2023). https://doi.org/10.1038/s41596-023-00867-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00867-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.