Abstract
Despite advances in automated liquid handling and microfluidics, preparing samples for RNA sequencing at scale generally requires expensive equipment, which is beyond the reach of many academic laboratories. Manual sample preparation remains a slow, expensive and error-prone process. Here, we describe a low-cost, semi-automated pipeline to extract cell-free RNA using one of two commercially available, inexpensive and open-source robotic systems: the Opentrons OT1.0 or OT2.0. Like many RNA isolation protocols, ours can be decomposed into three subparts: RNA extraction, DNA digestion and RNA cleaning and concentration. RT–qPCR data using a synthetic spike-in confirms comparable RNA quality to the gold standard, manual sample processing. The semi-automated pipeline also shows improvement in sample throughput (+12×), time spent (−11×), cost (−3×) and biohazardous waste produced (−4×) compared with its manual counterpart. This protocol enables cell-free RNA extraction from 96 samples simultaneously in 4.5 h; in practice, this dramatically improves the time to results, as we recently demonstrated. Importantly, any laboratory already has most of the parts required (manual pipette and corresponding tips and kits for RNA isolation, cleaning and concentration) to build a semi-automated sample processing pipeline of their own and would only need to purchase or three-dimensionally print a few extra parts (US$5.5 K–12 K in total). This pipeline is also generalizable for many nucleic acid extraction applications, thereby increasing the scale of studies, which can be performed in small research laboratories.
Key points
-
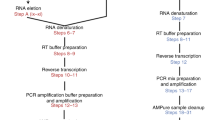
A protocol for setting up, calibrating and validating a semi-automated sample processing pipeline for cell-free RNA isolation from clinical samples using the Opentrons 1.0 or 2.0 open-source robotic platform.
-
The key benefits of this semi-automated system, as compared with manual extraction, include an increase in sample throughput by 12 fold. When compared with other available protocols for automated RNA extraction, this protocol provides a complementary option for larger sample volumes (>0.5 mL).
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Source data are provided with this paper. This file includes raw data to reproduce Fig. 3b–d and Fig. 4. The Supplementary Manual also includes instructions for how to build a system similar to the OT1 from scratch.
Code availability
All software, including the data analysis required to generate Figs. 3 and 4, are available at https://github.com/miramou/cfRNA_pipeline_automation (https://doi.org/10.5821/zenodo.7931552) (ref. 24) and https://github.com/miramou/cfRNA_pipeline (https://doi.org/10.5281/zenodo.7931554) (ref. 25).
References
Koh, W. et al. Noninvasive in vivo monitoring of tissue-specific global gene expression in humans. Proc. Natl Acad. Sci. USA 111, 7361–7366 (2014).
Fan, H. C. et al. Non-invasive prenatal measurement of the fetal genome. Nature 487, 320–324 (2012).
Lo, Y. M. D. et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2, 61ra91 (2010).
Tsang, J. C. H. et al. Integrative single-cell and cell-free plasma RNA transcriptomics elucidates placental cellular dynamics. Proc. Natl Acad. Sci. USA 114, E7786–E7795 (2017).
Pan, W. et al. Simultaneously monitoring immune response and microbial infections during pregnancy through plasma cfRNA sequencing. Clin. Chem. 63, 1695–1704 (2017).
Song, C.-X., Diao, J., Brunger, A. T. & Quake, S. R. Simultaneous single molecule epigenetic imaging of DNA methylation and hydroxymethylation. Proc. Natl Acad. Sci. USA 113, 4338–4343 (2016).
Crowley, E., Di Nicolantonio, F., Loupakis, F. & Bardelli, A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 10, 472–484 (2013).
Song, C.-X. et al. 5-Hydroxymethylcytosine signatures in cell-free DNA provide information about tumor types and stages. Cell Res. 27, 1231–1242 (2017).
Liu, M. C. et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31, 745–759 (2020).
Snyder, T. M., Khush, K. K., Valantine, H. A. & Quake, S. R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl Acad. Sci. USA 108, 6229–6234 (2011).
Sharon, E. et al. Quantification of transplant-derived circulating cell-free DNA in absence of a donor genotype. PLoS Comput. Biol. 13, e1005629 (2017).
Fan, H. C., Blumenfeld, Y. J., Chitkara, U., Hudgins, L. & Quake, S. R. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc. Natl Acad. Sci. USA 105, 16266–16271 (2008).
Ngo, T. T. M. et al. Noninvasive blood tests for fetal development predict gestational age and preterm delivery. Science 360, 1133–1136 (2018).
Moufarrej, M. N. et al. Early prediction of preeclampsia in pregnancy with cell-free RNA. Nature 602, 689–694 (2022).
Rasmussen, M. et al. RNA profiles reveal signatures of future health and disease in pregnancy. Nature 601, 422–427 (2022).
Camunas-Soler, J. et al. Predictive RNA profiles for early and very early spontaneous preterm birth. Am. J. Obstet. 227, 72.e1–72.e16 (2022).
Aghaeepour, N. et al. An immune clock of human pregnancy. Sci. Immunol. 2, eaan3946 (2017).
Aghaeepour, N. et al. A proteomic clock of human pregnancy. Am. J. Obstet. 218, 347.e1–347.e14 (2018).
Ghaemi, M. S. et al. Multiomics modeling of the immunome, transcriptome, microbiome, proteome and metabolome adaptations during human pregnancy. Bioinformatics 35, 95–103 (2019).
Tan, S. C. & Yiap, B. C. DNA, RNA, and protein extraction: the past and the present. Biomed Res. Int. 2009, 574398 (2009).
Lazaro-Perona, F. et al. Evaluation of two automated low-cost RNA extraction protocols for SARS-CoV-2 detection. PloS One 16, e0246302 (2021).
Mamanova, L. et al. High-throughput full-length single-cell RNA-seq automation. Nat. Protoc. 16, 2886–2915 (2021).
Moufarrej, M. N., Wong, R. J., Shaw, G. M., Stevenson, D. K. & Quake, S. R. Investigating pregnancy and its complications using circulating cell-free RNA in women’s blood during gestation. Front. Pediatr. 8, 605219 (2020).
Moufarrej, M. N., An inexpensive, semi-automated sample processing pipeline for cell-free RNA extraction, https://github.com/miramou/cfRNA_pipeline_automation, https://doi.org/10.5281/zenodo.7931552 (2023).
Moufarrej, M. N., An inexpensive, semi-automated sample processing pipeline for cell-free RNA extraction, https://github.com/miramou/cfrna_pipeline, https://doi.org/10.5281/zenodo.7931554 (2023).
Moufarrej, M. N., Early prediction of preeclampsia in pregnancy with cell-free RNA, https://github.com/miramou/pe_cfrna, https://doi.org/10.5281/zenodo.7931550 (2022).
Acknowledgements
We are grateful to both N. Neff and R. Sit for their sequencing expertise. We also thank B. Yu for helping brainstorm and providing indispensable advice around how to best use the Opentrons system. Figures 1 and 2D,E were created with BioRender.com. This work was supported by the Chan Zuckerberg Biohub. M.N.M. is supported by the Stanford Bio-X Bowes Fellowship.
Author information
Authors and Affiliations
Contributions
M.N.M. conceptualized and designed this protocol, and collected and analyzed the data in collaboration with S.R.Q. All authors contributed to the writing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.R.Q. is a founder, consultant and shareholder of Mirvie. M.N.M. is also a shareholder of Mirvie.
Peer review
Peer review information
Nature Protocols thanks Eleni Tomazou and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Moufarrej, M. N. et al. Nature 602, 689–694 (2022): https://doi.org/10.1038/s41586-022-04410-z
Moufarrej, M. (2020): https://youtu.be/g6RsSaNvSNA
Supplementary information
Supplementary Information
Supplementary Manual.
Source data
Source Data Figs. 3 and 4
Unprocessed RT–qPCR data for Figs. 3a–c and 4.
Rights and permissions
About this article
Cite this article
Moufarrej, M.N., Quake, S.R. An inexpensive semi-automated sample processing pipeline for cell-free RNA extraction. Nat Protoc 18, 2772–2793 (2023). https://doi.org/10.1038/s41596-023-00855-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-023-00855-2
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.