Abstract
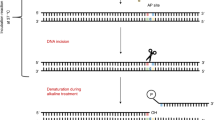
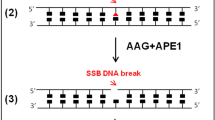
The comet assay is a versatile method to detect nuclear DNA damage in individual eukaryotic cells, from yeast to human. The types of damage detected encompass DNA strand breaks and alkali-labile sites (e.g., apurinic/apyrimidinic sites), alkylated and oxidized nucleobases, DNA–DNA crosslinks, UV-induced cyclobutane pyrimidine dimers and some chemically induced DNA adducts. Depending on the specimen type, there are important modifications to the comet assay protocol to avoid the formation of additional DNA damage during the processing of samples and to ensure sufficient sensitivity to detect differences in damage levels between sample groups. Various applications of the comet assay have been validated by research groups in academia, industry and regulatory agencies, and its strengths are highlighted by the adoption of the comet assay as an in vivo test for genotoxicity in animal organs by the Organisation for Economic Co-operation and Development. The present document includes a series of consensus protocols that describe the application of the comet assay to a wide variety of cell types, species and types of DNA damage, thereby demonstrating its versatility.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


















Similar content being viewed by others
Data availability
The majority of the data shown here as examples or anticipated results are available in the original papers. Figures 12 and 14–16 are theoretical results, which are inspired by unpublished work from the authors’ laboratories. Other supporting data are available upon reasonable request to the corresponding author.
References
Olive, P., Banáth, J. & Durand, R. Heterogeneity in radiation-induced DNA damage and repair in tumor and normal cells measured using the ‘comet’ assay. Radiat. Res. 122, 86–94 (1990).
Neri, M. et al. Worldwide interest in the comet assay: a bibliometric study. Mutagenesis 30, 155–163 (2015).
de Lapuente, J. et al. The comet assay and its applications in the field of ecotoxicology: a mature tool that continues to expand its perspectives. Front. Genet. 6, 180 (2015).
Gajski, G. et al. The comet assay in animal models: from bugs to whales—(Part 1 Invertebrates). Mutat. Res. Rev. Mutat. Res. 779, 82–113 (2019).
Gajski, G. et al. The comet assay in animal models: from bugs to whales—(Part 2 Vertebrates). Mutat. Res. Rev. Mutat. Res. 781, 130–164 (2019).
McKelvey-Martin, V. J. et al. The single cell gel electrophoresis assay (comet assay): a European review. Mutat. Res. Mol. Mech. Mutagen. 288, 47–63 (1993).
Tice, R. R. et al. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen. 35, 206–221 (2000).
OECD. Test No. 489: In Vivo Mammalian Alkaline Comet Assay (OECD Publishing, 2014).
ESCODD (European Standards Committee on Oxidative DNA Damage). Comparative analysis of baseline 8-oxo-7,8-dihydroguanine in mammalian cell DNA, by different methods in different laboratories: an approach to consensus. Carcinogenesis 23, 2129–2133 (2002).
ESCODD (European Standards Committee on Oxidative DNA Damage). Measurement of DNA oxidation in human cells by chromatographic and enzymic methods. Free Radic. Biol. Med. 34, 1089–1099 (2003).
Gedik, C. M. & Collins, A. Establishing the background level of base oxidation in human lymphocyte DNA: results of an interlaboratory validation study. FASEB J. 19, 82–84 (2005).
Møller, P., Moller, L., Godschalk, R. W. L. & Jones, G. D. D. Assessment and reduction of comet assay variation in relation to DNA damage: studies from the European Comet Assay Validation Group. Mutagenesis 25, 109–111 (2010).
Forchhammer, L. et al. Variation in the measurement of DNA damage by comet assay measured by the ECVAG inter-laboratory validation trial. Mutagenesis 25, 113–123 (2010).
Forchhammer, L. et al. Inter-laboratory variation in DNA damage using a standard comet assay protocol. Mutagenesis 27, 665–672 (2012).
Johansson, C. et al. An ECVAG trial on assessment of oxidative damage to DNA measured by the comet assay. Mutagenesis 25, 125–132 (2010).
Godschalk, R. W. L. et al. DNA-repair measurements by use of the modified comet assay: an inter-laboratory comparison within the European Comet Assay Validation Group (ECVAG). Mutat. Res. Toxicol. Environ. Mutagen. 757, 60–67 (2013).
Godschalk, R. W. L. et al. Variation of DNA damage levels in peripheral blood mononuclear cells isolated in different laboratories. Mutagenesis 29, 241–249 (2014).
Ersson, C. et al. An ECVAG inter-laboratory validation study of the comet assay: inter-laboratory and intra-laboratory variations of DNA strand breaks and FPG-sensitive sites in human mononuclear cells. Mutagenesis 28, 279–286 (2013).
Møller, P. et al. Potassium bromate as positive assay control for the Fpg-modified comet assay. Mutagenesis 35, 341–348 (2020).
Azqueta, A. et al. Application of the comet assay in human biomonitoring: an hCOMET perspective. Mutat. Res. Mutat. Res. 783, 108288 (2020).
Azqueta, A. et al. Technical recommendations to perform the alkaline standard and enzyme-modified comet assay in human biomonitoring studies. Mutat. Res. Toxicol. Environ. Mutagen. 843, 24–32 (2019).
Vodenkova, S. et al. An optimized comet-based in vitro DNA repair assay to assess base and nucleotide excision repair activity. Nat. Protoc. 15, 3844–3878 (2020).
Møller, P. et al. Minimum Information for Reporting on the Comet Assay (MIRCA): recommendations for describing comet assay procedures and results. Nat. Protoc. 15, 3817–3826 (2020).
Olive, P. L. & Banáth, J. P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 1, 23–29 (2006).
Ostling, O. & Johanson, K. J. Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochem. Biophys. Res. Commun. 123, 291–298 (1984).
Singh, N. P., McCoy, M. T., Tice, R. R. & Schneider, E. L. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175, 184–191 (1988).
Møller, P. The comet assay: ready for 30 more years. Mutagenesis 33, 1–7 (2018).
Olive, P. L., Wlodek, D. & Banáth, J. P. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 51, 4671–4676 (1991).
Shaposhnikov, S. et al. Twelve-gel slide format optimised for comet assay and fluorescent in situ hybridisation. Toxicol. Lett. 195, 31–34 (2010).
Gutzkow, K. B. et al. High-throughput comet assay using 96 minigels. Mutagenesis 28, 333–340 (2013).
Watson, C. et al. High-throughput screening platform for engineered nanoparticle-mediated genotoxicity using CometChip technology. ACS Nano 8, 2118–2133 (2014).
Collins, A. R. Measuring oxidative damage to DNA and its repair with the comet assay. Biochim. Biophys. Acta 1840, 794–800 (2014).
Collins, A. R. Investigating oxidative DNA damage and its repair using the comet assay. Mutat. Res. Mutat. Res. 681, 24–32 (2009).
Muruzabal, D., Collins, A. & Azqueta, A. The enzyme-modified comet assay: past, present and future. Food Chem. Toxicol. 147, 111865 (2021).
Wu, J. H. & Jones, N. J. Assessment of DNA interstrand crosslinks using the modified alkaline comet assay. Methods Mol. Biol. 817, 165–181 (2012).
Merk, O. & Speit, G. Detection of crosslinks with the comet assay in relationship to genotoxicity and cytotoxicity. Environ. Mol. Mutagen. 33, 167–172 (1999).
Spanswick, V. J., Hartley, J. M. & Hartley, J. A. in Drug–DNA Interaction Protocols. Methods in Molecular Biology (Methods and Protocols) (ed. Fox, K.) 267–282 (Humana Press, 2010).
Shaposhnikov, S., Frengen, E. & Collins, A. R. Increasing the resolution of the comet assay using fluorescent in situ hybridization—a review. Mutagenesis 24, 383–389 (2009).
Glei, M., Hovhannisyan, G. & Pool-Zobel, B. L. Use of comet–FISH in the study of DNA damage and repair: review. Mutat. Res. 681, 33–43 (2009).
Horváthová, E., Dusinská, M., Shaposhnikov, S. & Collins, A. R. DNA damage and repair measured in different genomic regions using the comet assay with fluorescent in situ hybridization. Mutagenesis 19, 269–276 (2004).
Spivak, G. Fluorescence in situ hybridization (FISH). Methods Mol. Biol. 659, 129–145 (2010).
Townsend, T. A., Parrish, M. C., Engelward, B. P. & Manjanatha, M. G. The development and validation of EpiComet-Chip, a modified high-throughput comet assay for the assessment of DNA methylation status. Environ. Mol. Mutagen. 58, 508–521 (2017).
Perotti, A., Rossi, V., Mutti, A. & Buschini, A. Methy-sens Comet assay and DNMTs transcriptional analysis as a combined approach in epigenotoxicology. Biomarkers 20, 64–70 (2015).
McKinnon, P. J. & Caldecott, K. W. DNA strand break repair and human genetic disease. Annu. Rev. Genomics Hum. Genet. 8, 37–55 (2007).
Chatterjee, N. & Walker, G. C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263 (2017).
Cheng, K., Cahill, D., Kasai, H., Nishimura, S. & Loeb, L. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G––T and A––C substitutions. J. Biol. Chem. 267, 166–172 (1992).
Collins, A. R., Duthie, S. J. & Dobson, V. L. Direct enzymic detection of endogenous oxidative base damage in human lymphocyte dna. Carcinogenesis 14, 1733–1735 (1993).
Dusinska, M. & Collins, A. Detection of oxidised purines and UV-induced photoproducts in DNA of single cells, by inclusion of lesion-specific enzymes in the comet assay. Altern. Lab. Anim. 24, 405–411 (1996).
Smith, C. C., O’Donovan, M. R. & Martin, E. A. hOGG1 recognizes oxidative damage using the comet assay with greater specificity than FPG or ENDOIII. Mutagenesis 21, 185–190 (2006).
Evans, M. D. et al. Detection of purine lesions in cellular DNA using single cell gel electrophoresis with Fpg protein. Biochem. Soc. Trans. 23, 434S (1995).
Muruzabal, D., Langie, S. A. S., Pourrut, B. & Azqueta, A. The enzyme-modified comet assay: enzyme incubation step in 2 vs 12-gels/slide systems. Mutat. Res. Toxicol. Environ. Mutagen. 845, 402981 (2019).
Collins, A., Dusinská, M. & Horská, A. Detection of alkylation damage in human lymphocyte DNA with the comet assay. Acta Biochim. Pol. 48, 11–14 (2001).
Hašplová, K. et al. DNA alkylation lesions and their repair in human cells: modification of the comet assay with 3-methyladenine DNA glycosylase (AlkD). Toxicol. Lett. 208, 76–81 (2012).
Muruzabal, D. et al. Novel approach for the detection of alkylated bases using the enzyme-modified comet assay. Toxicol. Lett. 330, 108–117 (2020).
Connor, T. R. O. Purification and characterization of human 3-methyladenine-DNA glycosylase. Nucleic Acids Res. 21, 5561–5569 (1993).
Lee, C.-Y. I. et al. Recognition and processing of a new repertoire of DNA substrates by human 3-methyladenine DNA glycosylase (AAG). Biochemistry 48, 1850–1861 (2009).
Azqueta, A., Arbillaga, L., Lopez de Cerain, A. & Collins, A. Enhancing the sensitivity of the comet assay as a genotoxicity test, by combining it with bacterial repair enzyme FPG. Mutagenesis 28, 271–277 (2013).
Hansen, S. H. et al. Using the comet assay and lysis conditions to characterize DNA lesions from the acrylamide metabolite glycidamide. Mutagenesis 33, 31–39 (2018).
Speit, G., Schütz, P., Bonzheim, I., Trenz, K. & Hoffmann, H. Sensitivity of the FPG protein towards alkylation damage in the comet assay. Toxicol. Lett. 146, 151–158 (2004).
Noll, D. M., Mason, T. M. & Miller, P. S. Formation and repair of interstrand cross-links in DNA. Chem. Rev. 106, 277–301 (2006).
Folmer, V., Soares, J. C. M., Gabriel, D. & Rocha, J. B. T. A high fat diet inhibits δ-aminolevulinate dehydratase and increases lipid peroxidation in mice (Mus musculus). J. Nutr. 133, 2165–2170 (2003).
Ljunggren, B. Severe phototoxic burn following celery ingestion. Arch. Dermatol. 126, 1334–1336 (1990).
Bennetts, L. E. et al. Impact of estrogenic compounds on DNA integrity in human spermatozoa: evidence for cross-linking and redox cycling activities. Mutat. Res. Mol. Mech. Mutagen. 641, 1–11 (2008).
Dextraze, M.-E., Gantchev, T., Girouard, S. & Hunting, D. DNA interstrand cross-links induced by ionizing radiation: an unsung lesion. Mutat. Res. Rev. Mutat. Res. 704, 101–107 (2010).
Olive, P. L. & Banáth, J. P. Sizing highly fragmented DNA in individual apoptotic cells using the comet assay and a DNA crosslinking agent. Exp. Cell Res. 221, 19–26 (1995).
Collins, A. R. et al. UV-sensitive rodent mutant cell lines of complementation groups 6 and 8 differ phenotypically from their human counterparts. Environ. Mol. Mutagen. 29, 152–160 (1997).
Collins, A. DNA repair in ultraviolet-irradiated HeLa cells is disrupted by aphidicolin. Biochim. Biophys. Acta Gene Struct. Expr. 741, 341–347 (1983).
Gedik, C. M., Ewen, S. W. B. & Collins, A. R. Single-cell gel electrophoresis applied to the analysis of UV-C damage and its repair in human cells. Int. J. Radiat. Biol. 62, 313–320 (1992).
Baranovskiy, A. G. et al. Structural basis for inhibition of DNA replication by aphidicolin. Nucleic Acids Res. 42, 14013–14021 (2014).
Cheng, C. H. & Kuchta, R. D. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry 32, 8568–8574 (1993).
Goscin, L. P. & Byrnes, J. J. DNA polymerase delta: one polypeptide, two activities. Biochemistry 21, 2513–2518 (1982).
Bausinger, J., Schütz, P., Piberger, A. L. & Speit, G. Further characterization of benzo[a]pyrene diol-epoxide (BPDE)-induced comet assay effects. Mutagenesis 31, 161–169 (2016).
Vande Loock, K., Decordier, I., Ciardelli, R., Haumont, D. & Kirsch-Volders, M. An aphidicolin-block nucleotide excision repair assay measuring DNA incision and repair capacity. Mutagenesis 25, 25–32 (2010).
Ngo, L. P. et al. Sensitive CometChip assay for screening potentially carcinogenic DNA adducts by trapping DNA repair intermediates. Nucleic Acids Res. 48, e13 (2020).
Azqueta, A. et al. A comparative performance test of standard, medium- and high-throughput comet assays. Toxicol. Vitr. 27, 768–773 (2013).
Guilherme, S., Santos, M. A., Barroso, C., Gaivão, I. & Pacheco, M. Differential genotoxicity of Roundup® formulation and its constituents in blood cells of fish (Anguilla anguilla): considerations on chemical interactions and DNA damaging mechanisms. Ecotoxicology 21, 1381–1390 (2012).
Guilherme, S., Santos, M. A., Gaivão, I. & Pacheco, M. Are DNA-damaging effects induced by herbicide formulations (Roundup® and Garlon®) in fish transient and reversible upon cessation of exposure? Aquat. Toxicol. 155, 213–221 (2014).
Brunborg, G. et al. High throughput sample processing and automated scoring. Front. Genet. 5, 373 (2014).
McNamee, J., McLean, J., Ferrarotto, C. & Bellier, P. Comet assay: rapid processing of multiple samples. Mutat. Res. Toxicol. Environ. Mutagen. 466, 63–69 (2000).
Perdry, H. et al. Validation of Gelbond® high-throughput alkaline and Fpg-modified comet assay using a linear mixed model. Environ. Mol. Mutagen. 59, 595–602 (2018).
Enciso, J. M. et al. Standardisation of the in vitro comet assay: influence of lysis time and lysis solution composition on the detection of DNA damage induced by X-rays. Mutagenesis 33, 25–30 (2018).
Wood, D. K., Weingeist, D. M., Bhatia, S. N. & Engelward, B. P. Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl Acad. Sci. USA 107, 10008–10013 (2010).
Weingeist, D. M. et al. Single-cell microarray enables high-throughput evaluation of DNA double-strand breaks and DNA repair inhibitors. Cell Cycle 12, 907–915 (2013).
Ge, J. et al. Micropatterned comet assay enables high throughput and sensitive DNA damage quantification. Mutagenesis 30, 11–19 (2015).
Ge, J. et al. Standard fluorescent imaging of live cells is highly genotoxic. Cytom. Part A 83A, 552–560 (2013).
Seo, J.-E. et al. Quantitative comparison of in vitro genotoxicity between metabolically competent HepaRG cells and HepG2 cells using the high-throughput high-content CometChip assay. Arch. Toxicol. 93, 1433–1448 (2019).
Mutamba, J. T. et al. XRCC1 and base excision repair balance in response to nitric oxide. DNA Repair 10, 1282–1293 (2011).
Chao, C., Ngo, L. P. & Engelward, B. P. SpheroidChip: patterned agarose microwell compartments harboring HepG2 spheroids are compatible with genotoxicity testing. ACS Biomater. Sci. Eng. 6, 2427–2439 (2020).
Chao, C. & Engelward, B. P. Applications of CometChip for environmental health studies. Chem. Res. Toxicol. 33, 1528–1538 (2020).
Karbaschi, M. & Cooke, M. S. Novel method for the high-throughput processing of slides for the comet assay. Sci. Rep. 4, 7200 (2015).
Lewies, A., Van Dyk, E., Wentzel, J. F. & Pretorius, P. J. Using a medium-throughput comet assay to evaluate the global DNA methylation status of single cells. Front. Genet. 5, 215 (2014).
Wentzel, J. F. et al. Assessing the DNA methylation status of single cells with the comet assay. Anal. Biochem. 400, 190–194 (2010).
Pogribny, I., Yi, P. & James, S. J. A sensitive new method for rapid detection of abnormal methylation patterns in global DNA and within CpG islands. Biochem. Biophys. Res. Commun. 262, 624–628 (1999).
Mohsen, K., Johansson, S. & Ekström, T. J. Using LUMA: a luminometric-based assay for global DNA-methylation. Epigenetics 1, 46–49 (2006).
Gowher, H., Leismann, O. & Jeltsch, A. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 19, 6918–6923 (2000).
Zhou, Y., Bui, T., Auckland, L. D. & Williams, C. G. Undermethylated DNA as a source of microsatellites from a conifer genome. Genome 45, 91–99 (2002).
Adamczyk, J. et al. Affected chromosome homeostasis and genomic instability of clonal yeast cultures. Curr. Genet. 62, 405–418 (2016).
Lewinska, A., Miedziak, B. & Wnuk, M. Assessment of yeast chromosome XII instability: single chromosome comet assay. Fungal Genet. Biol. 63, 9–16 (2014).
Krol, K. et al. Lack of G1/S control destabilizes the yeast genome via replication stress-induced DSBs and illegitimate recombination. J. Cell Sci. 131, jcs226480 (2018).
Cecchini, M. J., Amiri, M. & Dick, F. A. Analysis of cell cycle position in mammalian cells. J. Vis. Exp. 3491 (2012).
Nagar, S., Hanley-Bowdoin, L. & Robertson, D. Host DNA replication is induced by geminivirus infection of differentiated plant cells. Plant Cell 14, 2995–3007 (2002).
Mórocz, M., Gali, H., Raskó, I., Downes, C. S. & Haracska, L. Single cell analysis of human RAD18-dependent DNA post-replication repair by alkaline bromodeoxyuridine comet assay. PLoS ONE 8, e70391 (2013).
McGlynn, A. P., Wasson, G., O’Connor, J., McKelvey-Martin, V. J. & Downes, C. S. The bromodeoxyuridine comet assay: detection of maturation of recently replicated DNA in individual cells. Cancer Res. 59, 5912–5916 (1999).
McGlynn, A. P. et al. Detection of replicative integrity in small colonic biopsies using the BrdUrd comet assay. Br. J. Cancer 88, 895–901 (2003).
Guo, J., Hanawalt, P. C. & Spivak, G. Comet–FISH with strand-specific probes reveals transcription-coupled repair of 8-oxoGuanine in human cells. Nucleic Acids Res. 41, 7700–7712 (2013).
Mladinic, M., Zeljezic, D., Shaposhnikov, S. A. & Collins, A. R. The use of FISH–comet to detect c-Myc and TP 53 damage in extended-term lymphocyte cultures treated with terbuthylazine and carbofuran. Toxicol. Lett. 211, 62–69 (2012).
Azevedo, F., Marques, F., Fokt, H., Oliveira, R. & Johansson, B. Measuring oxidative DNA damage and DNA repair using the yeast comet assay. Yeast 28, 55–61 (2011).
Oliveira, R. & Johansson, B. in DNA Repair Protocols, Methods in Molecular Biology (ed. Bjergbæk, L.) 101–109 (Humana Press, 2012).
Santos, C. L. V., Pourrut, B. & Ferreira de Oliveira, J. M. P. The use of comet assay in plant toxicology: recent advances. Front. Genet. 6, 216 (2015).
Dhawan, A., Bajpayee, M. & Parmar, D. Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol. Toxicol. 25, 5–32 (2009).
Jha, A. N. Ecotoxicological applications and significance of the comet assay. Mutagenesis 23, 207–221 (2008).
Ghosh, M., Ghosh, I., Godderis, L., Hoet, P. & Mukherjee, A. Genotoxicity of engineered nanoparticles in higher plants. Mutat. Res. Toxicol. Environ. Mutagen. 842, 132–145 (2019).
Lanier, C., Manier, N., Cuny, D. & Deram, A. The comet assay in higher terrestrial plant model: review and evolutionary trends. Environ. Pollut. 207, 6–20 (2015).
Bajpayee, M., Kumar, A. & Dhawan, A. in Genotoxicity Assessment: Methods and Protocols (Methods in Molecular Biology series) (eds Dhawan, A. & Bajpayee. M.) Vol 2031, 237–257 (Humana Press, 2019).
Pedron, J. et al. Novel 8-nitroquinolin-2(1H)-ones as NTR-bioactivated antikinetoplastid molecules: Synthesis, electrochemical and SAR study. Eur. J. Med. Chem. 155, 135–152 (2018).
Le Hégarat, L. et al. Performance of comet and micronucleus assays in metabolic competent HepaRG cells to predict in vivo genotoxicity. Toxicol. Sci. 138, 300–309 (2014).
Cowie, H. et al. Suitability of human and mammalian cells of different origin for the assessment of genotoxicity of metal and polymeric engineered nanoparticles. Nanotoxicology 9, 57–65 (2015).
Naik, U. C., Das, M. T., Sauran, S. & Thakur, I. S. Assessment of in vitro cyto/genotoxicity of sequentially treated electroplating effluent on the human hepatocarcinoma HuH-7 cell line. Mutat. Res. Toxicol. Environ. Mutagen. 762, 9–16 (2014).
Waldherr, M. et al. Use of HuH6 and other human-derived hepatoma lines for the detection of genotoxins: a new hope for laboratory animals? Arch. Toxicol. 92, 921–934 (2018).
Kruszewski, M. et al. Comet assay in neural cells as a tool to monitor DNA damage induced by chemical or physical factors relevant to environmental and occupational exposure. Mutat. Res. Toxicol. Environ. Mutagen. 845, 402990 (2019).
Borm, P. J. A., Fowler, P. & Kirkland, D. An updated review of the genotoxicity of respirable crystalline silica. Part. Fibre Toxicol. 15, 23 (2018).
Bankoglu, E. E., Kodandaraman, G. & Stopper, H. A systematic review of the use of the alkaline comet assay for genotoxicity studies in human colon-derived cells. Mutat. Res. Toxicol. Environ. Mutagen. 845, 402976 (2019).
Møller, P. et al. Applications of the comet assay in particle toxicology: air pollution and engineered nanomaterials exposure. Mutagenesis 30, 67–83 (2015).
Wischermann, K., Boukamp, P. & Schmezer, P. Improved alkaline comet assay protocol for adherent HaCaT keratinocytes to study UVA-induced DNA damage. Mutat. Res. Toxicol. Environ. Mutagen. 630, 122–128 (2007).
García-Rodríguez, A., Vila, L., Cortés, C., Hernández, A. & Marcos, R. Effects of differently shaped TiO2NPs (nanospheres, nanorods and nanowires) on the in vitro model (Caco-2/HT29) of the intestinal barrier. Part. Fibre Toxicol. 15, 33 (2018).
Domenech, J., Hernández, A., Demir, E., Marcos, R. & Cortés, C. Interactions of graphene oxide and graphene nanoplatelets with the in vitro Caco-2/HT29 model of intestinal barrier. Sci. Rep. 10, 2793 (2020).
Ventura, C. et al. Cytotoxicity and genotoxicity of MWCNT-7 and crocidolite: assessment in alveolar epithelial cells versus their coculture with monocyte-derived macrophages. Nanotoxicology 14, 479–503 (2020).
Ventura, C., Lourenço, A. F., Sousa-Uva, A., Ferreira, P. J. T. & Silva, M. J. Evaluating the genotoxicity of cellulose nanofibrils in a co-culture of human lung epithelial cells and monocyte-derived macrophages. Toxicol. Lett. 291, 173–183 (2018).
Jantzen, K. et al. Oxidative damage to DNA by diesel exhaust particle exposure in co-cultures of human lung epithelial cells and macrophages. Mutagenesis 27, 693–701 (2012).
Machado, A. R. T. et al. Cytotoxic, genotoxic, and oxidative stress-inducing effect of an l-amino acid oxidase isolated from Bothrops jararacussu venom in a co-culture model of HepG2 and HUVEC cells. Int. J. Biol. Macromol. 127, 425–432 (2019).
Žegura, B. & Filipič, M. The application of the comet assay in fish cell lines. Mutat. Res. Toxicol. Environ. Mutagen. 842, 72–84 (2019).
Canedo, A. & Rocha, T. L. Zebrafish (Danio rerio) using as model for genotoxicity and DNA repair assessments: Historical review, current status and trends. Sci. Total Environ. 762, 144084 (2021).
Reeves, J. F., Davies, S. J., Dodd, N. J. F. & Jha, A. N. Hydroxyl radicals (OH) are associated with titanium dioxide (TiO2) nanoparticle-induced cytotoxicity and oxidative DNA damage in fish cells. Mutat. Res. Mol. Mech. Mutagen. 640, 113–122 (2008).
Fuchs, R. et al. Modification of the alkaline comet assay with human mesenchymal stem cells. Cell Biol. Int. 36, 113–117 (2012).
Tomc, J. et al. Adipose tissue stem cell-derived hepatic progenies as an in vitro model for genotoxicity testing. Arch. Toxicol. 92, 1893–1903 (2018).
Garcia, A. L. H. et al. Fluorosilicic acid induces DNA damage and oxidative stress in bone marrow mesenchymal stem cells. Mutat. Res. Toxicol. Environ. Mutagen. 861–862, 503297 (2021).
Hiemstra, P. S., Grootaers, G., van der Does, A. M., Krul, C. A. M. & Kooter, I. M. Human lung epithelial cell cultures for analysis of inhaled toxicants: lessons learned and future directions. Toxicol. Vitr. 47, 137–146 (2018).
Štampar, M., Tomc, J., Filipič, M. & Žegura, B. Development of in vitro 3D cell model from hepatocellular carcinoma (HepG2) cell line and its application for genotoxicity testing. Arch. Toxicol. 93, 3321–3333 (2019).
Mišík, M. et al. Use of human derived liver cells for the detection of genotoxins in comet assays. Mutat. Res. Toxicol. Environ. Mutagen. 845, 402995 (2019).
Pfuhler, S. et al. Use of in vitro 3D tissue models in genotoxicity testing: strategic fit, validation status and way forward. Report of the working group from the 7th International Workshop on Genotoxicity Testing (IWGT). Mutat. Res. Toxicol. Environ. Mutagen. 850–851, 503135 (2020).
Pfuhler, S. et al. A tiered approach to the use of alternatives to animal testing for the safety assessment of cosmetics: genotoxicity. A COLIPA analysis. Regul. Toxicol. Pharmacol. 57, 315–324 (2010).
Pfuhler, S. et al. The Cosmetics Europe strategy for animal-free genotoxicity testing: project status up-date. Toxicol. Vitr. 28, 18–23 (2014).
Reisinger, K. et al. Validation of the 3D Skin Comet assay using full thickness skin models: transferability and reproducibility. Mutat. Res. Toxicol. Environ. Mutagen. 827, 27–41 (2018).
Reus, A. A. et al. Comet assay in reconstructed 3D human epidermal skin models–investigation of intra- and inter-laboratory reproducibility with coded chemicals. Mutagenesis 28, 709–720 (2013).
Pfuhler, S. et al. Validation of the 3D reconstructed human skin comet assay, an animal-free alternative for following-up positive results from standard in vitro genotoxicity assays. Mutagenesis 36, 19–35 (2021).
Elje, E. et al. The comet assay applied to HepG2 liver spheroids. Mutat. Res. Toxicol. Environ. Mutagen. 845, 403033 (2019).
Mandon, M., Huet, S., Dubreil, E., Fessard, V. & Le Hégarat, L. Three-dimensional HepaRG spheroids as a liver model to study human genotoxicity in vitro with the single cell gel electrophoresis assay. Sci. Rep. 9, 10548 (2019).
Elje, E. et al. Hepato(geno)toxicity assessment of nanoparticles in a HepG2 liver spheroid model. Nanomaterials 10, 545 (2020).
Štampar, M. et al. Hepatocellular carcinoma (HepG2/C3A) cell-based 3D model for genotoxicity testing of chemicals. Sci. Total Environ. 755, 143255 (2021).
Kooter, I. M. et al. Cellular effects in an in vitro human 3D cellular airway model and A549/BEAS-2B in vitro cell cultures following air exposure to cerium oxide particles at an air–liquid interface. Appl. Vitr. Toxicol. 2, 56–66 (2016).
Strähle, U. et al. Zebrafish embryos as an alternative to animal experiments—a commentary on the definition of the onset of protected life stages in animal welfare regulations. Reprod. Toxicol. 33, 128–132 (2012).
Kelly, J. R. & Benson, S. A. Inconsistent ethical regulation of larval zebrafish in research. J. Fish. Biol. 97, 324–327 (2020).
Kosmehl, T. et al. A novel contact assay for testing genotoxicity of chemicals and whole sediments in zebrafish embryos. Environ. Toxicol. Chem. 25, 2097–2106 (2006).
Deregowska, A. et al. Shifts in rDNA levels act as a genome buffer promoting chromosome homeostasis. Cell Cycle 14, 3475–3487 (2015).
Cerda, H., Hofsten, B. & Johanson, K. in Proceedings of the Workshop on Recent Advances on Detection of Irradiated Food (eds. Leonardi, M., Bessiardo, J. & Raffi, J.) 401–405 (Commission of the European Communities, 1993).
Koppen, G. & Verschaeve, L. The alkaline comet test on plant cells: a new genotoxicity test for DNA strand breaks in Vicia faba root cells. Mutat. Res. Mutagen. Relat. Subj. 360, 193–200 (1996).
Einset, J. & Collins, A. R. DNA repair after X-irradiation: lessons from plants. Mutagenesis 30, 45–50 (2015).
Gichner, T., Znidar, I., Wagner, E. D. & Plewa, M. J. in The Comet Assay in Toxicology (eds. Dhawan, A. & Anderson, D.) 98–119 (Royal Society of Chemistry, 2009).
Pellegri, V., Gorbi, G. & Buschini, A. Comet assay on Daphnia magna in eco-genotoxicity testing. Aquat. Toxicol. 155, 261–268 (2014).
Parrella, A., Lavorgna, M., Criscuolo, E., Russo, C. & Isidori, M. Eco-genotoxicity of six anticancer drugs using comet assay in daphnids. J. Hazard. Mater. 286, 573–580 (2015).
Russo, C., Kundi, M., Lavorgna, M., Parrella, A. & Isidori, M. Benzalkonium chloride and anticancer drugs in binary mixtures: reproductive toxicity and genotoxicity in the freshwater crustacean Ceriodaphnia dubia. Arch. Environ. Contam. Toxicol. 74, 546–556 (2018).
Lavorgna, M., Russo, C., D’Abrosca, B., Parrella, A. & Isidori, M. Toxicity and genotoxicity of the quaternary ammonium compound benzalkonium chloride (BAC) using Daphnia magna and Ceriodaphnia dubia as model systems. Environ. Pollut. 210, 34–39 (2016).
Kundi, M. et al. Prediction and assessment of ecogenotoxicity of antineoplastic drugs in binary mixtures. Environ. Sci. Pollut. Res. 23, 14771–14779 (2016).
Sario, S., Silva, A. M. & Gaivão, I. Titanium dioxide nanoparticles: toxicity and genotoxicity in Drosophila melanogaster (SMART eye-spot test and comet assay in neuroblasts). Mutat. Res. Toxicol. Environ. Mutagen. 831, 19–23 (2018).
Gaivão, I. & Sierra, L. M. Drosophila comet assay: insights, uses, and future perspectives. Front. Genet. 5, 304 (2014).
Marques, A. et al. Comparative genoprotection ability of wild-harvested vs. aqua-cultured Ulva rigida coupled with phytochemical profiling. Eur. J. Phycol. 56, 105–118 (2021).
Bilbao, C., Ferreiro, J. A., Comendador, M. A. & Sierra, L. M. Influence of mus201 and mus308 mutations of Drosophila melanogaster on the genotoxicity of model chemicals in somatic cells in vivo measured with the comet assay. Mutat. Res. Mol. Mech. Mutagen. 503, 11–19 (2002).
Mukhopadhyay, I., Chowdhuri, D. K., Bajpayee, M. & Dhawan, A. Evaluation of in vivo genotoxicity of cypermethrin in Drosophila melanogaster using the alkaline comet assay. Mutagenesis 19, 85–90 (2004).
Siddique, H. R., Chowdhuri, D. K., Saxena, D. K. & Dhawan, A. Validation of Drosophila melanogaster as an in vivo model for genotoxicity assessment using modified alkaline comet assay. Mutagenesis 20, 285–290 (2005).
Sharma, A., Shukla, A. K., Mishra, M. & Chowdhuri, D. K. Validation and application of Drosophila melanogaster as an in vivo model for the detection of double strand breaks by neutral comet assay. Mutat. Res. Toxicol. Environ. Mutagen. 721, 142–146 (2011).
Ribeiro, I. P. & Gaivão, I. Efeito genotóxico do etanol em neuroblastos de Drosophila melanogaster. Rev. Port. Saúde. Pública 28, 199–204 (2010).
Brennan, L. J., Haukedal, J. A., Earle, J. C., Keddie, B. & Harris, H. L. Disruption of redox homeostasis leads to oxidative DNA damage in spermatocytes of Wolbachia-infected Drosophila simulans. Insect Mol. Biol. 21, 510–520 (2012).
Verma, A., Sengupta, S. & Lakhotia, S. C. DNApol-ϵ gene is indispensable for the survival and growth of Drosophila melanogaster. Genesis 50, 86–101 (2012).
Carmona, E. R., Guecheva, T. N., Creus, A. & Marcos, R. Proposal of an in vivo comet assay using haemocytes of Drosophila melanogaster. Environ. Mol. Mutagen. 52, 165–169 (2011).
Augustyniak, M., Gladysz, M. & Dziewięcka, M. The Comet assay in insects—Status, prospects and benefits for science. Mutat. Res. Rev. Mutat. Res. 767, 67–76 (2016).
Kadhim, M. A. Methodologies for monitoring the genetic effects of mutagens and carcinogens accumulated in the body of marine mussels. Rev. Aquat. Sci. 2, 83–107 (1990).
Prego-Faraldo, M. V., Valdiglesias, V., Laffon, B., Eirín-López, J. M. & Méndez, J. In vitro analysis of early genotoxic and cytotoxic effects of okadaic acid in different cell types of the mussel Mytilus galloprovincialis. J. Toxicol. Environ. Heal. Part A 78, 814–824 (2015).
Jha, A. N., Dogra, Y., Turner, A. & Millward, G. E. Impact of low doses of tritium on the marine mussel, Mytilus edulis: genotoxic effects and tissue-specific bioconcentration. Mutat. Res. Toxicol. Environ. Mutagen. 586, 47–57 (2005).
Nacci, D. E., Cayula, S. & Jackim, E. Detection of DNA damage in individual cells from marine organisms using the single cell gel assay. Aquat. Toxicol. 35, 197–210 (1996).
Steinert, S. A. Contribution of apoptosis to observed DNA damage in mussel cells. Mar. Environ. Res. 42, 253–259 (1996).
Wilson, J., Pascoe, P., Parry, J. & Dixon, D. Evaluation of the comet assay as a method for the detection of DNA damage in the cells of a marine invertebrate, Mytilus edulis L. (Mollusca: Pelecypoda). Mutat. Res. Mol. Mech. Mutagen. 399, 87–95 (1998).
Mitchelmore, C. & Chipman, J. DNA strand breakage in aquatic organisms and the potential value of the comet assay in environmental monitoring. Mutat. Res. Mol. Mech. Mutagen. 399, 135–147 (1998).
Frenzilli, G., Nigro, M. & Lyons, B. The comet assay for the evaluation of genotoxic impact in aquatic environments. Mutat. Res. Rev. Mutat. Res. 681, 80–92 (2009).
Gentile, L., Cebrià, F. & Bartscherer, K. The planarian flatworm: an in vivo model for stem cell biology and nervous system regeneration. Dis. Model. Mech. 4, 12–19 (2011).
Guecheva, T., Henriques, J. A. & Erdtmann, B. Genotoxic effects of copper sulphate in freshwater planarian in vivo, studied with the single-cell gel test (comet assay). Mutat. Res. Toxicol. Environ. Mutagen. 497, 19–27 (2001).
Stevens, A.-S. et al. Planarians customize their stem cell responses following genotoxic stress as a function of exposure time and regenerative state. Toxicol. Sci. 162, 251–263 (2018).
Peiris, T. H. et al. Regional signals in the planarian body guide stem cell fate in the presence of genomic instability. Development 143, 1697–1709 (2016).
Thiruvalluvan, M., Barghouth, P. G., Tsur, A., Broday, L. & Oviedo, N. J. SUMOylation controls stem cell proliferation and regional cell death through Hedgehog signaling in planarians. Cell. Mol. Life Sci. 75, 1285–1301 (2018).
Yin, S. et al. SmedOB1 is required for planarian homeostasis and regeneration. Sci. Rep. 6, 34013 (2016).
Eyambe, G. S., Goven, A. J., Fitzpatrick, L. C., Venables, B. J. & Cooper, E. L. A non-invasive technique for sequential collection of earthworm (Lumbricus terrestris) leukocytes during subchronic immunotoxicity studies. Lab. Anim. 25, 61–67 (1991).
Verschaeve, L. & Gilles, J. Single cell gel electrophoresis assay in the earthworm for the detection of genotoxic compounds in soils. Bull. Environ. Contam. Toxicol. 54, 112–119 (1995).
Reinecke, S. A. & Reinecke, A. J. The comet assay as biomarker of heavy metal genotoxicity in earthworms. Arch. Environ. Contam. Toxicol. 46, 208–215 (2004).
Lourenço, J. I. et al. Genotoxic endpoints in the earthworms sub-lethal assay to evaluate natural soils contaminated by metals and radionuclides. J. Hazard. Mater. 186, 788–795 (2011).
Lladó, S., Solanas, A. M., de Lapuente, J., Borràs, M. & Viñas, M. A diversified approach to evaluate biostimulation and bioaugmentation strategies for heavy-oil-contaminated soil. Sci. Total Environ. 435–436, 262–269 (2012).
Sforzini, S. et al. Genotoxicity assessment in Eisenia andrei coelomocytes: a study of the induction of DNA damage and micronuclei in earthworms exposed to B[a]P- and TCDD-spiked soils. Mutat. Res. Toxicol. Environ. Mutagen. 746, 35–41 (2012).
Saez, G. et al. Genotoxic and oxidative responses in coelomocytes of Eisenia fetida and Hediste diversicolor exposed to lipid-coated CdSe/ZnS quantum dots and CdCl2. Environ. Toxicol. 30, 918–926 (2015).
Ramadass, K. et al. Earthworm comet assay for assessing the risk of weathered petroleum hydrocarbon contaminated soils: need to look further than target contaminants. Arch. Environ. Contam. Toxicol. 71, 561–571 (2016).
Jiang, X. et al. Toxicological effects of polystyrene microplastics on earthworm (Eisenia fetida). Environ. Pollut. 259, 113896 (2020).
Ralph, S., Petras, M., Pandrangi, R. & Vrzoc, M. Alkaline single-cell gel (comet) assay and genotoxicity monitoring using two species of tadpoles. Environ. Mol. Mutagen. 28, 112–120 (1996).
Cotelle S, F. J. Comet assay in genetic ecotoxicology: a review. Environ. Mol. Mutagen. 34, 246–255 (1999).
Pandrangi, R., Petras, M., Ralph, S. & Vrzoc, M. Alkaline single cell gel (comet) assay and genotoxicity monitoring using bullheads and carp. Environ. Mol. Mutagen. 26, 345–356 (1995).
Pereira, V. et al. Marine macroalgae as a dietary source of genoprotection in gilthead seabream (Sparus aurata) against endogenous and exogenous challenges. Comp. Biochem. Physiol. Part C. Toxicol. Pharmacol. 219, 12–24 (2019).
Burlinson, B. et al. Fourth International Workgroup on Genotoxicity testing: results of the in vivo comet assay workgroup. Mutat. Res. Toxicol. Environ. Mutagen. 627, 31–35 (2007).
Hartmann, A. et al. Recommendations for conducting the in vivo alkaline comet assay. Mutagenesis 18, 45–51 (2003).
Uno, Y. et al. JaCVAM-organized international validation study of the in vivo rodent alkaline comet assay for the detection of genotoxic carcinogens: II. Summary of definitive validation study results. Mutat. Res. Toxicol. Environ. Mutagen. 786–788, 45–76 (2015).
Morita, T. et al. The JaCVAM international validation study on the in vivo comet assay: selection of test chemicals. Mutat. Res. Toxicol. Environ. Mutagen. 786–788, 14–44 (2015).
Sasaki, Y. F. et al. The comet assay with multiple mouse organs: comparison of comet assay results and carcinogenicity with 208 chemicals selected from the IARC Monographs and U.S. NTP Carcinogenicity Database. Crit. Rev. Toxicol. 30, 629–799 (2000).
Uno, Y. et al. JaCVAM-organized international validation study of the in vivo rodent alkaline comet assay for the detection of genotoxic carcinogens: I. Summary of pre-validation study results. Mutat. Res. Toxicol. Environ. Mutagen. 786–788, 3–13 (2015).
Pool-Zobel, B. L. et al. Assessment of genotoxic effects by Lindane. Food Chem. Toxicol. 31, 271–283 (1993).
Giovannelli, L., Decorosi, F., Dolara, P. & Pulvirenti, L. Vulnerability to DNA damage in the aging rat substantia nigra: a study with the comet assay. Brain Res. 969, 244–247 (2003).
Vestergaard, S., Loft, S. & Møller, P. Role of inducible nitrogen oxide synthase in benzene-induced oxidative DNA damage in the bone marrow of mice. Free Radic. Biol. Med. 32, 481–484 (2002).
Doak, S. H. & Dusinska, M. NanoGenotoxicology: present and the future. Mutagenesis 32, 1–4 (2017).
Risom, L., Møller, P., Kristjansen, P., Loft, S. & Vogel, U. X-ray-induced oxidative stress: DNA damage and gene expression of HO-1, ERCC1 and OGG1 in mouse lung. Free Radic. Res. 37, 957–966 (2003).
Schupp, N., Schmid, U., Heidland, A. & Stopper, H. New Approaches for the Treatment of Genomic Damage in End-Stage Renal Disease. J. Ren. Nutr. 18, 127–133 (2008).
Gunasekarana, V. A comprehensive review on clinical applications of comet assay. J. Clin. Diagnostic Res. 9, GE01–GE05 (2015).
Gajski, G. et al. Analysis of health-related biomarkers between vegetarians and non-vegetarians: a multi-biomarker approach. J. Funct. Foods 48, 643–653 (2018).
Fagundes, G. E. et al. Vitamin D3 as adjuvant in the treatment of type 2 diabetes mellitus: modulation of genomic and biochemical instability. Mutagenesis 34, 135–145 (2019).
Macan, T. P. et al. Brazil nut prevents oxidative DNA damage in type 2 diabetes patients. Drug Chem. Toxicol. 45, 1066–1072 (2022).
Kuchařová, M. et al. Comet assay and its use for evaluating oxidative DNA damage in some pathological states. Physiol. Res. 68, 1–15 (2019).
Møller, P., Stopper, H. & Collins, A. R. Measurement of DNA damage with the comet assay in high-prevalence diseases: current status and future directions. Mutagenesis 35, 5–18 (2020).
Gomolka, M. et al. Age-dependent differences in DNA damage after in vitro CT exposure. Int. J. Radiat. Biol. 94, 272–281 (2018).
Ziegler, B. L. et al. Short-term effects of early-acting and multilineage hematopoietic growth factors on the repair and proliferation of irradiated pure cord blood (CB) CD34 hematopoietic progenitor cells. Int. J. Radiat. Oncol. 40, 1193–1203 (1998).
Wyatt, N. et al. In vitro susceptibilities in lymphocytes from mothers and cord blood to the monofunctional alkylating agent EMS. Mutagenesis 22, 123–127 (2007).
Wang, L. et al. Characterization of placenta-derived mesenchymal stem cells cultured in autologous human cord blood serum. Mol. Med. Rep. 6, 760–766 (2012).
Menon, R. et al. Senescence of primary amniotic cells via oxidative DNA damage. PLoS ONE 8, e83416 (2013).
Želježić, D., Herceg Romanić, S., Klinčić, D., Matek Sarić, M. & Letinić, J. G. Persistent organochlorine pollutants in placentas sampled from women in Croatia and an evaluation of their DNA damaging potential in vitro. Arch. Environ. Contam. Toxicol. 74, 284–291 (2018).
Chao, M.-R. et al. Biomarkers of nucleic acid oxidation—a summary state-of-the-art. Redox Biol. 42, 101872 (2021).
Møller, P. et al. Collection and storage of human white blood cells for analysis of DNA damage and repair activity using the comet assay in molecular epidemiology studies. Mutagenesis 36, 193–212 (2021).
Collins, A. R. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 26, 249–261 (2004).
Ladeira, C. & Smajdova, L. The use of genotoxicity biomarkers in molecular epidemiology: applications in environmental, occupational and dietary studies. AIMS Genet. 4, 166–191 (2017).
Collins, A. R. et al. Controlling variation in the comet assay. Front. Genet. 5, 359 (2014).
Valverde, M. & Rojas, E. Environmental and occupational biomonitoring using the comet assay. Mutat. Res. Rev. Mutat. Res. 681, 93–109 (2009).
Gajski, G. et al. Application of the comet assay for the evaluation of DNA damage from frozen human whole blood samples: implications for human biomonitoring. Toxicol. Lett. 319, 58–65 (2020).
Gajski, G., Gerić, M., Oreščanin, V. & Garaj-Vrhovac, V. Cytogenetic status of healthy children assessed with the alkaline comet assay and the cytokinesis-block micronucleus cytome assay. Mutat. Res. Toxicol. Environ. Mutagen. 750, 55–62 (2013).
Garaj-Vrhovac, V. et al. Assessment of cytogenetic damage and oxidative stress in personnel occupationally exposed to the pulsed microwave radiation of marine radar equipment. Int. J. Hyg. Environ. Health 214, 59–65 (2011).
Gerić, M., Gajski, G., Oreščanin, V. & Garaj-Vrhovac, V. Seasonal variations as predictive factors of the comet assay parameters: a retrospective study. Mutagenesis 33, 53–60 (2018).
Azqueta, A., Shaposhnikov, S. & Collins, A. R in The Comet Assay in Toxicology (eds Dhawan, A. & Anderson, D.) Ch. 2, 57–78 (RSC Publishing, 2009).
Gerić, M. et al. Cytogenetic status and oxidative stress parameters in patients with thyroid diseases. Mutat. Res. Toxicol. Environ. Mutagen. 810, 22–29 (2016).
Milić, M. et al. Alkaline comet assay results on fresh and one-year frozen whole blood in small volume without cryo-protection in a group of people with different health status. Mutat. Res. Toxicol. Environ. Mutagen. 843, 3–10 (2019).
Bankoglu, E. E. et al. Reduction of DNA damage in peripheral lymphocytes of obese patients after bariatric surgery-mediated weight loss. Mutagenesis 33, 61–67 (2018).
Milković, Đ. et al. Primary DNA damage assessed with the comet assay and comparison to the absorbed dose of diagnostic X-rays in children. Int. J. Toxicol. 28, 405–416 (2009).
Milić, M. et al. The hCOMET project: international database comparison of results with the comet assay in human biomonitoring. Baseline frequency of DNA damage and effect of main confounders. Mutat. Res. Rev. Mutat. Res. 787, 108371 (2021).
Giovannelli, L., Pitozzi, V., Riolo, S. & Dolara, P. Measurement of DNA breaks and oxidative damage in polymorphonuclear and mononuclear white blood cells: a novel approach using the comet assay. Mutat. Res. Toxicol. Environ. Mutagen. 538, 71–80 (2003).
Martelli-Palomino, G. et al. DNA damage increase in peripheral neutrophils from patients with rheumatoid arthritis is associated with the disease activity and the presence of shared epitope. Clin. Exp. Rheumatol. 35, 247–254 (2017).
McConnell, J. R., Crockard, A. D., Cairns, A. P. & Bell, A. L. Neutrophils from systemic lupus erythematosus patients demonstrate increased nuclear DNA damage. Clin. Exp. Rheumatol. 20, 653–660 (2003).
Wong, C. H. et al. Sevoflurane-induced oxidative stress and cellular injury in human peripheral polymorphonuclear neutrophils. Food Chem. Toxicol. 44, 1399–1407 (2006).
Zielińska-Przyjemska, M., Olejnik, A., Dobrowolska-Zachwieja, A., Łuczak, M. & Baer-Dubowska, W. DNA damage and apoptosis in blood neutrophils of inflammatory bowel disease patients and in Caco-2 cells in vitro exposed to betanin. Postepy Hig. Med. Dosw. 70, 265–271 (2016).
Sul, D. et al. Single strand DNA breaks in T- and B-lymphocytes and granulocytes in workers exposed to benzene. Toxicol. Lett. 134, 87–95 (2002).
Sul, D. et al. DNA damage in T- and B-lymphocytes and granulocytes in emission inspection and incineration workers exposed to polycyclic aromatic hydrocarbons. Mutat. Res. Toxicol. Environ. Mutagen. 538, 109–119 (2003).
Marino, M. et al. Impact of 12-month cryopreservation on endogenous DNA damage in whole blood and isolated mononuclear cells evaluated by the comet assay. Sci. Rep. 11, 363 (2021).
Al-Salmani, K. et al. Simplified method for the collection, storage, and comet assay analysis of DNA damage in whole blood. Free Radic. Biol. Med. 51, 719–725 (2011).
Bøhn, S. K., Vebraite, V., Shaposhnikov, S. & Collins, A. R. Isolation of leukocytes from frozen buffy coat for comet assay analysis of DNA damage. Mutat. Res. Toxicol. Environ. Mutagen. 843, 18–23 (2019).
Decordier, I. et al. Genetic susceptibility of newborn daughters to oxidative stress. Toxicol. Lett. 172, 68–84 (2007).
Knudsen, L. E. & Hansen, Å. M. Biomarkers of intermediate endpoints in environmental and occupational health. Int. J. Hyg. Environ. Health 210, 461–470 (2007).
Norishadkam, M., Andishmand, S., Zavar reza, J., Zare Sakhvidi, M. J. & Hachesoo, V. R. Oxidative stress and DNA damage in the cord blood of preterm infants. Mutat. Res. Toxicol. Environ. Mutagen. 824, 20–24 (2017).
Gelaleti, R. B., Damasceno, D. C., Santos, D. P., Calderon, I. M. P. & Rudge, M. V. C. Increased DNA damage is related to maternal blood glucose levels in the offspring of women with diabetes and mild gestational hyperglycemia. Reprod. Sci. 23, 318–323 (2016).
Oßwald, K., Mittas, A., Glei, M. & Pool-Zobel, B. L. New revival of an old biomarker: characterisation of buccal cells and determination of genetic damage in the isolated fraction of viable leucocytes. Mutat. Res. Rev. Mutat. Res. 544, 321–329 (2003).
Feretti, D. et al. Monitoring air pollution effects on children for supporting public health policy: the protocol of the prospective cohort MAPEC study. BMJ Open 4, e006096 (2014).
Zani, C. et al. Comet test in saliva leukocytes of pre-school children exposed to air pollution in North Italy: the Respira study. Int. J. Environ. Res. Public Health 17, 3276 (2020).
Rojas, E., Lorenzo, Y., Haug, K., Nicolaissen, B. & Valverde, M. Epithelial cells as alternative human biomatrices for comet assay. Front. Genet. 5, 386 (2014).
Souto, E. B. et al. Ocular cell lines and genotoxicity assessment. Int. J. Environ. Res. Public Health 17, 2046 (2020).
Rojas, E., Valverde, M., Sordo, M. & Ostrosky-Wegman, P. DNA damage in exfoliated buccal cells of smokers assessed by the single cell gel electrophoresis assay. Mutat. Res. Toxicol. 370, 115–120 (1996).
Sánchez-Alarcón, J., Milić, M., Gómez-Arroyo, S., Montiel-González, J. M. R. & Valencia-Quintana, R. in Environmental Health Risk - Hazardous Factors to Living Species (eds Larramendy, M. L. & Soloneski, S.) (IntechOpen, 2016); https://doi.org/10.5772/62760
Valverde, M. et al. DNA damage in leukocytes and buccal and nasal epithelial cells of individuals exposed to air pollution in Mexico City. Environ. Mol. Mutagen. 30, 147–152 (1997).
Calderón-Garcidueñas, L. et al. 8-Hydroxy-2′-deoxyguanosine, a major mutagenic oxidative DNA lesion, and DNA strand breaks in nasal respiratory epithelium of children exposed to urban pollution. Environ. Health Perspect. 107, 469–474 (1999).
Godderis, L. et al. Influence of genetic polymorphisms on biomarkers of exposure and genotoxic effects in styrene-exposed workers. Environ. Mol. Mutagen. 44, 293–303 (2004).
Koreck, A. et al. Effects of intranasal phototherapy on nasal mucosa in patients with allergic rhinitis. J. Photochem. Photobiol. B Biol. 89, 163–169 (2007).
Akkaş, H., Aydın, E., Türkoğlu-Babakurban, S., Yurtcu, E. & Yılmaz-Özbek, Ö. Effect of mometasone furoate nasal spray on the DNA of nasal mucosal cells. Turk. J. Med. Sci. 48, 339–345 (2018).
Anderson, D. Factors that contribute to biomarker responses in humans including a study in individuals taking vitamin C supplementation. Mutat. Res. Mol. Mech. Mutagen. 480–481, 337–347 (2001).
Baumeister, P., Huebner, T., Reiter, M., Schwenk-Zieger, S. & Harréus, U. Reduction of oxidative DNA fragmentation by ascorbic acid, zinc and N-acetylcysteine in nasal mucosa tissue cultures. Anticancer Res. 29, 4571–4574 (2009).
Koehler, C. et al. Aspects of nitrogen dioxide toxicity in environmental urban concentrations in human nasal epithelium. Toxicol. Appl. Pharmacol. 245, 219–225 (2010).
Reiter, M., Rupp, K., Baumeister, P., Zieger, S. & Harréus, U. Antioxidant effects of quercetin and coenzyme Q10 in mini organ cultures of human nasal mucosa cells. Anticancer Res. 29, 33–39 (2009).
Mrowicka, M., Zielinska-Blizniewska, H., Milonski, J., Olszewski, J. & Majsterek, I. Evaluation of oxidative DNA damage and antioxidant defense in patients with nasal polyps. Redox Rep. 20, 177–183 (2015).
Zhang, J. et al. DNA damage in lens epithelial cells and peripheral lymphocytes from age-related cataract patients. Ophthalmic Res. 51, 124–128 (2014).
Rojas, E. et al. Evaluation of DNA damage in exfoliated tear duct epithelial cells from individuals exposed to air pollution assessed by single cell gel electrophoresis assay. Mutat. Res. Toxicol. Environ. Mutagen. 468, 11–17 (2000).
Gajski, G. et al. Application of the comet assay for the evaluation of DNA damage in mature sperm. Mutat. Res. Rev. Mutat. Res. 788, 108398 (2021).
Sipinen, V. et al. In vitro evaluation of baseline and induced DNA damage in human sperm exposed to benzo[a]pyrene or its metabolite benzo[a]pyrene-7,8-diol-9,10-epoxide, using the comet assay. Mutagenesis 25, 417–425 (2010).
Hamilton, T. R., dos, S. & Assumpção, M. E. O. D. Sperm DNA fragmentation: causes and identification. Zygote 28, 1–8 (2020).
Agarwal, A., Barbăroșie, C., Ambar, R. & Finelli, R. The impact of single- and double-strand DNA breaks in human spermatozoa on assisted reproduction. Int. J. Mol. Sci. 21, 3882 (2020).
Sugihara, A., Van Avermaete, F., Roelant, E., Punjabi, U. & De Neubourg, D. The role of sperm DNA fragmentation testing in predicting intra-uterine insemination outcome: a systematic review and meta-analysis. Eur. J. Obstet. Gynecol. Reprod. Biol. 244, 8–15 (2020).
Simon, L., Aston, K. I., Emery, B. R., Hotaling, J. & Carrell, D. T. Sperm DNA damage output parameters measured by the alkaline comet assay and their importance. Andrologia 49, e12608 (2017).
Nicopoullos, J. et al. Novel use of COMET parameters of sperm DNA damage may increase its utility to diagnose male infertility and predict live births following both IVF and ICSI. Hum. Reprod. 34, 1915–1923 (2019).
Simon, L. et al. Clinical significance of sperm DNA damage in assisted reproduction outcome. Hum. Reprod. 25, 1594–1608 (2010).
Albert, O., Reintsch, W. E., Chan, P. & Robaire, B. HT-COMET: a novel automated approach for high throughput assessment of human sperm chromatin quality. Hum. Reprod. 31, 938–946 (2016).
Fry, R. C., Bangma, J., Szilagyi, J. & Rager, J. E. Developing novel in vitro methods for the risk assessment of developmental and placental toxicants in the environment. Toxicol. Appl. Pharmacol. 378, 114635 (2019).
Vähäkangas, K. et al. in Biomarkers in Toxicology 325–360 (Elsevier, 2014).
Kohn, K. W., Ewig, R. A. G., Erickson, L. C. & Zwelling, L. A. in DNA Repair: A Laboratory Manual of Research Procedures (eds. Friedberg, E. C. & Hanawalt, P. C.) 379–401 (Marcel Dekker Inc, 1981).
Ahnström, G. & Erixon, K. in DNA Repair: A Laboratory Manual of Research Procedures (eds. Friedberg, E. C. & Hanawalt, P. C.) 403–418 (Marcel Dekker Inc, 1981).
Gedik, C. M., Boyle, S. P., Wood, S. G., Vaughan, N. J. & Collins, A. R. Oxidative stress in humans: validation of biomarkers of DNA damage. Carcinogenesis 23, 1441–1446 (2002).
Pflaum, M., Will, O. & Epe, B. Determination of steady-state levels of oxidative DNA base modifications in mammalian cells by means of repair endonucleases. Carcinogenesis 18, 2225–2231 (1997).
Fujiwara, H. & Ito, M. Nonisotopic cytosine extension assay: a highly sensitive method to evaluate CpG island methylation in the whole genome. Anal. Biochem. 307, 386–389 (2002).
Azqueta, A. & Collins, A. The essential comet assay: a comprehensive guide to measuring DNA damage and repair. Arch. Toxicol. 87, 949–968 (2013).
Azqueta, A. et al. The influence of scoring method on variability in results obtained with the comet assay. Mutagenesis 26, 393–399 (2011).
Møller, P. et al. On the search for an intelligible comet assay descriptor. Front. Genet. 17, 217 (2014).
Collins, A. et al. The comet assay as a tool for human biomonitoring studies: the ComNet Project. Mutat. Res. Rev. Mutat. Res. 759, 27–39 (2014).
Azqueta, A., Gutzkow, K. B., Brunborg, G. & Collins, A. R. Towards a more reliable comet assay: optimising agarose concentration, unwinding time and electrophoresis conditions. Mutat. Res. Toxicol. Environ. Mutagen. 724, 41–45 (2011).
Ersson, C. & Moller, L. The effects on DNA migration of altering parameters in the comet assay protocol such as agarose density, electrophoresis conditions and durations of the enzyme or the alkaline treatments. Mutagenesis 26, 689–695 (2011).
Kumaravel, T. S., Vilhar, B., Faux, S. P. & Jha, A. N. Comet assay measurements: a perspective. Cell Biol. Toxicol. 25, 53–64 (2009).
Kumaravel, T. S. & Jha, A. N. Reliable comet assay measurements for detecting DNA damage induced by ionising radiation and chemicals. Mutat. Res. 605, 7–16 (2006).
Langie, S. A. S., Azqueta, A. & Collins, A. R. The comet assay: past, present, and future. Front. Genet. 6, 266 (2015).
Kohn, K. W., Erickson, L. C. & Ewig, R. A. G. Fractionation of DNA from mammalian cells by alkaline elution. Biochemistry 15, 4629–4637 (1976).
García, O. et al. Sensitivity and variability of visual scoring in the comet assay. Mutat. Res. Mol. Mech. Mutagen. 556, 25–34 (2004).
Bonassi, S. et al. DNA damage in circulating leukocytes measured with the comet assay may predict the risk of death. Sci. Rep. 11, 16793 (2021).
Anderson, D., Yu, T.-W. & McGregor, D. B. Comet assay responses as indicators of carcinogen exposure. Mutagenesis 13, 539–555 (1998).
Bowen, D. E. et al. Evaluation of a multi-endpoint assay in rats, combining the bone-marrow micronucleus test, the comet assay and the flow-cytometric peripheral blood micronucleus test. Mutat. Res. Toxicol. Environ. Mutagen. 722, 7–19 (2011).
Kirkland, D. et al. A comparison of transgenic rodent mutation and in vivo comet assay responses for 91 chemicals. Mutat. Res. Toxicol. Environ. Mutagen. 839, 21–35 (2019).
Akor-Dewu, M. B. et al. Leucocytes isolated from simply frozen whole blood can be used in human biomonitoring for DNA damage measurement with the comet assay. Cell Biochem. Funct. 32, 299–302 (2014).
Ladeira, C., Koppen, G., Scavone, F. & Giovannelli, L. The comet assay for human biomonitoring: effect of cryopreservation on DNA damage in different blood cell preparations. Mutat. Res. Toxicol. Environ. Mutagen. 843, 11–17 (2019).
Hininger, I. et al. Assessment of DNA damage by comet assay on frozen total blood: method and evaluation in smokers and non-smokers. Mutat. Res. Toxicol. Environ. Mutagen. 558, 75–80 (2004).
Koppen, G. et al. The comet assay in human biomonitoring: cryopreservation of whole blood and comparison with isolated mononuclear cells. Mutagenesis 33, 41–47 (2018).
Bankoglu, E. E. et al. Effect of cryopreservation on DNA damage and DNA repair activity in human blood samples in the comet assay. Arch. Toxicol. 95, 1831–1841 (2021).
Azqueta, A., Enciso, J. M., Pastor, L., López de Cerain, A. & Vettorazzi, A. Applying the comet assay to fresh vs frozen animal solid tissues: a technical approach. Food Chem. Toxicol. 132, 110671 (2019).
Møller, P. et al. Searching for assay controls for the Fpg- and hOGG1-modified comet assay. Mutagenesis 33, 9–19 (2018).
Pfuhler, S., Downs, T. R., Allemang, A. J., Shan, Y. & Crosby, M. E. Weak silica nanomaterial-induced genotoxicity can be explained by indirect DNA damage as shown by the OGG1-modified comet assay and genomic analysis. Mutagenesis 32, 5–12 (2017).
Speit, G., Trenz, K., Schütz, P., Rothfuß, A. & Merk, O. The influence of temperature during alkaline treatment and electrophoresis on results obtained with the comet assay. Toxicol. Lett. 110, 73–78 (1999).
Sirota, N. P. et al. Some causes of inter-laboratory variation in the results of comet assay. Mutat. Res. Toxicol. Environ. Mutagen. 770, 16–22 (2014).
Singh, N. P., Stephens, R. E. & Schneider, E. L. Modifications of alkaline microgel electrophoresis for sensitive detection of DNA damage. Int. J. Radiat. Biol. 66, 23–28 (1994).
Møller, P., Loft, S., Lundby, C. & Olsen, N. V. Acute hypoxia and hypoxic exercise induce DNA strand breaks and oxidative DNA damage in humans. FASEB J. 15, 1181–1186 (2001).
Ji, Y., Karbaschi, M. & Cooke, M. S. Mycoplasma infection of cultured cells induces oxidative stress and attenuates cellular base excision repair activity. Mutat. Res. Toxicol. Environ. Mutagen. 845, 403054 (2019).
Zarcone, M. C. et al. Cellular response of mucociliary differentiated primary bronchial epithelial cells to diesel exhaust. Am. J. Physiol. Cell. Mol. Physiol. 311, L111–L123 (2016).
Eleršek, T., Plazar, J. & Filipič, M. A method for the assessment of DNA damage in individual, one day old, zebrafish embryo (Danio rerio), without prior cell isolation. Toxicol. Vitr. 27, 2156–2159 (2013).
Martins, C. & Costa, P. M. Technical updates to the comet assay in vivo for assessing DNA damage in zebrafish embryos from fresh and frozen cell suspensions. Zebrafish 17, 220–228 (2020).
Koppen, G. & Angelis, K. J. Repair of X-ray induced DNA damage measured by the comet assay in roots of Vicia faba. Environ. Mol. Mutagen. 32, 281–285 (1998).
Koppen, G., Toncelli, L., Triest, L. & Verschaeve, L. The comet assay: a tool to study alteration of DNA integrity in developing plant leaves. Mech. Ageing Dev. 110, 13–24 (1999).
Jackson, P. et al. Validation of freezing tissues and cells for analysis of DNA strand break levels by comet assay. Mutagenesis 28, 699–707 (2013).
Belpaeme, K., Cooreman, K. & Kirsch-Volders, M. Development and validation of the in vivo alkaline comet assay for detecting genomic damage in marine flatfish. Mutat. Res. Toxicol. Environ. Mutagen. 415, 167–184 (1998).
Braz, M. G. & Karahalil, B. Genotoxicity of anesthetics evaluated in vivo (animals). Biomed. Res. Int. 2015, 1–8 (2015).
Fernández-Bertólez, N., Azqueta, A., Pásaro, E., Laffon, B. & Valdiglesias, V. Salivary leucocytes as suitable biomatrix for the comet assay in human biomonitoring studies. Arch. Toxicol. 95, 2179–2187 (2021).
Russo, C., Acito, M., Fatigoni, C., Villarini, M. & Moretti, M. B-comet assay (comet assay on buccal cells) for the evaluation of primary DNA damage in human biomonitoring studies. Int. J. Environ. Res. Public Health 17, 9234 (2020).
Fortoul, T. I. et al. Single-cell gel electrophoresis assay of nasal epithelium and leukocytes from asthmatic and nonasthmatic subjects in Mexico City. Arch. Environ. Health 58, 348–352 (2003).
Osnes-Ringen, Ø. et al. DNA damage in lens epithelium of cataract patients in vivo and ex vivo. Acta Ophthalmol. 91, 652–656 (2013).
Olsen, A.-K. et al. Highly efficient base excision repair (BER) in human and rat male germ cells. Nucleic Acids Res. 29, 1781–1790 (2001).
Lorenzo, Y., Costa, S., Collins, A. R. & Azqueta, A. The comet assay, DNA damage, DNA repair and cytotoxicity: hedgehogs are not always dead. Mutagenesis 28, 427–432 (2013).
Henderson, L., Wolfreys, A., Fedyk, J., Bourner, C. & Windebank, S. The ability of the comet assay to discriminate between genotoxins and cytotoxins. Mutagenesis 13, 89–94 (1998).
Pfuhler, S. & Wolf, H. U. Detection of DNA-crosslinking agents with the alkaline comet assay. Environ. Mol. Mutagen. 27, 196–201 (1996).
Møller, P., Wallin, H., Dybdahl, M., Frentz, G. & Nexø, B. A. Psoriasis patients with basal cell carcinoma have more repair-mediated DNA strand-breaks after UVC damage in lymphocytes than psoriasis patients without basal cell carcinoma. Cancer Lett. 151, 187–192 (2000).
Møller, P., Knudsen, L. E., Loft, S. & Wallin, H. The comet assay as a rapid test in biomonitoring occupational exposure to DNA-damaging agents and effect of confounding factors. Cancer Epidemiol. Biomark. Prev. 9, 1005–1015 (2000).
Jensen, A. et al. Influence of the OGG1 Ser326Cys polymorphism on oxidatively damaged DNA and repair activity. Free Radic. Biol. Med. 52, 118–125 (2012).
Shaposhnikov, S. A., Salenko, V. B., Brunborg, G., Nygren, J. & Collins, A. R. Single-cell gel electrophoresis (the comet assay): loops or fragments? Electrophoresis 29, 3005–3012 (2008).
Vesterdal, L. K. et al. Accumulation of lipids and oxidatively damaged DNA in hepatocytes exposed to particles. Toxicol. Appl. Pharmacol. 274, 350–360 (2014).
Acknowledgements
We thank the hCOMET project (COST Action, CA 15132) for support. A. Azqueta thanks the Ministry of Science and Innovation (AGL2015-70640-R and PID2020-115348RB-I00) of the Spanish Government. S.G. thanks the national funds (OE), through FCT—Fundação para a Ciência e a Tecnologia (IP, in the scope of the framework contract foreseen in the numbers 4, 5 and 6 of the article 23, of the Decree-Law 57/2016, of 29 August, changed by Law 57/2017, of 19 July) for personal support. V.M.d.A. thanks the National Council of Technological and Scientific Development (CNPq—304203/2018-1) for personal support. D.M. thanks the program ‘Ayudas para la formación de profesorado universitario (FPU)’ of the Spanish Government for the predoctoral grant received. N.O. thanks the NIEHS Superfund Research Program ES ES027707 for personal support. J.S.-S. thanks the Government of Navarra for the predoctoral grant received. V.V. thanks the Ministerio de Educación, Cultura y Deporte (‘Beatriz Galindo’ program, BEAGAL18/00142) of the Spanish Government for personal support. M.S.C. acknowledges personal support from the National Institute of Environmental Health Sciences of the National Institutes of Health under award number: 1R41ES030274-01. This paper reflects the views of the authors and does not necessarily reflect those of the US Food and Drug Administration or the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
P.M., S.V., S.A.S.L., K.B.G., M.S.C., B.E., J.F.W. and S.S. designed figures; P.M. provided anticipated results; A.C., G.G., P.M., S.V., S.A.S.L. and A. Azqueta drafted the paper and revised the manuscript; all other co-authors contributed to the Materials and Procedure sections; A.L.d.C., E.B.-R., F.J.v.S., M.S.C., S.C. and S.K. thoroughly reviewed the manuscript before submission; S.A.S.L. and A. Azqueta managed the manuscript preparation; all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Vilena Kašuba and Bryant Nelson for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Gajski, G. et al. Mutat. Res. Rev. Mutat. Res. 779, 82–113 (2019): https://doi.org/10.1016/j.mrrev.2019.02.003
Gajski, G. et al. Mutat. Res. Rev. Mutat. Res. 781, 130–164 (2019): https://doi.org/10.1016/j.mrrev.2019.04.002
Azqueta, A. et al. Mutat. Res. Rev. Mutat. Res. 783, 108288 (2020): https://doi.org/10.1016/j.mrrev.2019.108288
Gajski, G. et al. Mutat. Res. Rev. Mutat. Res. 788, 108398 (2021): https://doi.org/10.1016/j.mrrev.2021.108398
Supplementary information
Supplementary Information
Supplementary Protocols 1–13.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Collins, A., Møller, P., Gajski, G. et al. Measuring DNA modifications with the comet assay: a compendium of protocols. Nat Protoc 18, 929–989 (2023). https://doi.org/10.1038/s41596-022-00754-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00754-y
This article is cited by
-
Copper toxicity on Eisenia fetida in a vineyard soil: a combined study with standard tests, genotoxicity assessment and gut metagenomic analysis
Environmental Science and Pollution Research (2024)
-
Assessment of toxicity, genotoxicity and oxidative stress in Fejervarya limnocharis exposed to tributyltin
Environmental Science and Pollution Research (2024)
-
Electrophoresis, a transport technology that transitioned from moving boundary method to zone method
European Biophysics Journal (2024)
-
Exposure of piperlongumine attenuates stemness and epithelial to mesenchymal transition phenotype with more potent anti-metastatic activity in SOX9 deficient human lung cancer cells
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Do hairdressers comprise a high risk group for genotoxicity? A systematic review
International Archives of Occupational and Environmental Health (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.