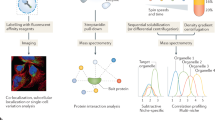
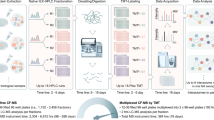
Abstract
The molecular functions of a protein are defined by its inherent properties in relation to its environment and interaction network. Within a cell, this environment and network are defined by the subcellular location of the protein. Consequently, it is crucial to know the localization of a protein to fully understand its functions. Recently, we have developed a mass spectrometry– (MS) and bioinformatics-based pipeline to generate a proteome-wide resource for protein subcellular localization across multiple human cancer cell lines (www.subcellbarcode.org). Here, we present a detailed wet-lab protocol spanning from subcellular fractionation to MS-sample preparation and analysis. A key feature of this protocol is that it includes all generated cell fractions without discarding any material during the fractionation process. We also describe the subsequent quantitative MS-data analysis, machine learning–based classification, differential localization analysis and visualization of the output. For broad applicability, we evaluated the pipeline by using MS data generated by two different peptide pre-fractionation approaches, namely high-resolution isoelectric focusing and high-pH reverse-phase fractionation, as well as direct analysis without pre-fractionation by using long-gradient liquid chromatography-MS. Moreover, an R package covering the dry-lab part of the method was developed and made available through Bioconductor. The method is straightforward and robust, and the entire protocol, from cell harvest to classification output, can be performed within 1–2 weeks. The protocol enables accurate classification of proteins to 15 compartments and 4 neighborhoods, visualization of the output data and differential localization analysis including treatment-induced protein relocalization, condition-dependent localization or cell type–specific localization. The SubCellBarCode package is freely available at https://bioconductor.org/packages/devel/bioc/html/SubCellBarCode.html.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The MS proteomics data for the analysis of the HeLa cell line have been deposited to the ProteomeXchange Consortium via the jPOST partner repository with the dataset identifier PXD022533. The SubCellBarCode R package and manual are freely available through the Bioconductor repository (https://doi.org/10.18129/B9.bioc.SubCellBarCode). The MS proteomics data for the previous analysis of five different cell lines have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006895. All described processed datasets can be easily accessed, explored and visualized at the SubCellBarCode portal (https://www.subcellbarcode.org). Source data are provided with this paper.
Code availability
The SubCellBarCode pipeline used in this protocol is available at https://github.com/TanerArslan/SubCellBarCode/ and can also be found in Bioconductor (https://bioconductor.org/packages/release/bioc/html/SubCellBarCode.html).
References
Heald, R. & Cohen-Fix, O. Morphology and function of membrane-bound organelles. Curr. Opin. Cell Biol. 26, 79–86 (2014).
Bauer, N. C., Doetsch, P. W. & Corbett, A. H. Mechanisms regulating protein localization. Traffic 16, 1039–1061 (2015).
Wang, A. J., Han, Y., Jia, N., Chen, P. & Minden, M. D. NPM1c impedes CTCF functions through cytoplasmic mislocalization in acute myeloid leukemia. Leukemia 34, 1278–1290 (2020).
Dansen, T. B. & Burgering, B. M. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 18, 421–429 (2008).
Guardia, C. M., De Pace, R., Mattera, R. & Bonifacino, J. S. Neuronal functions of adaptor complexes involved in protein sorting. Curr. Opin. Neurobiol. 51, 103–110 (2018).
De Matteis, M. A. & Luini, A. Mendelian disorders of membrane trafficking. N. Engl. J. Med. 365, 927–938 (2011).
Thul, P. J. et al. A subcellular map of the human proteome. Science 356, eaal3321 (2017).
Schnell, U., Dijk, F., Sjollema, K. A. & Giepmans, B. N. Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods 9, 152–158 (2012).
Stadler, C. et al. Immunofluorescence and fluorescent-protein tagging show high correlation for protein localization in mammalian cells. Nat. Methods 10, 315–323 (2013).
Andersen, J. S. et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426, 570–574 (2003).
Foster, L. J. et al. A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 (2006).
Liu, X., Salokas, K., Weldatsadik, R. G., Gawriyski, L. & Varjosalo, M. Combined proximity labeling and affinity purification-mass spectrometry workflow for mapping and visualizing protein interaction networks. Nat. Protoc. 15, 3182–3211 (2020).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012).
Gatto, L., Breckels, L. M., Wieczorek, S., Burger, T. & Lilley, K. S. Mass-spectrometry-based spatial proteomics data analysis using pRoloc and pRolocdata. Bioinformatics 30, 1322–1324 (2014).
Christoforou, A. et al. A draft map of the mouse pluripotent stem cell spatial proteome. Nat. Commun. 7, 8992 (2016).
Itzhak, D. N., Tyanova, S., Cox, J. & Borner, G. H. Global, quantitative and dynamic mapping of protein subcellular localization. eLife 5, e16950 (2016).
Itzhak, D. N. et al. A mass spectrometry-based approach for mapping protein subcellular localization reveals the spatial proteome of mouse primary neurons. Cell Rep. 20, 2706–2718 (2017).
Geladaki, A. et al. Combining LOPIT with differential ultracentrifugation for high-resolution spatial proteomics. Nat. Commun. 10, 331 (2019).
Orre, L. M. et al. SubCellBarCode: proteome-wide mapping of protein localization and relocalization. Mol. Cell 73, 166–182.e7 (2019).
Joshi, R. N. et al. TcellSubC: an atlas of the subcellular proteome of human T cells. Front. Immunol. 10, 2708 (2019).
Stenström, L. et al. Mapping the nucleolar proteome reveals a spatiotemporal organization related to intrinsic protein disorder. Mol. Syst. Biol. 16, e9469 (2020).
Herr, P. et al. Cell cycle profiling reveals protein oscillation, phosphorylation, and localization dynamics. Mol. Cell. Proteom. 19, 608–623 (2020).
Moll, T., Tebb, G., Surana, U., Robitsch, H. & Nasmyth, K. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5. Cell 66, 743–758 (1991).
Du, J. X., Bialkowska, A. B., McConnell, B. B. & Yang, V. W. SUMOylation regulates nuclear localization of Kruppel-like factor 5. J. Biol. Chem. 283, 31991–32002 (2008).
Wang, M. & Casey, P. J. Protein prenylation: unique fats make their mark on biology. Nat. Rev. Mol. Cell Biol. 17, 110–122 (2016).
Mertins, P. et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography–mass spectrometry. Nat. Protoc. 13, 1632–1661 (2018).
Christopher, J. A. et al. Subcellular proteomics. Nat. Rev. Methods Prim. 1, 32 (2021).
Lundberg, E. & Borner, G. H. H. Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 20, 285–302 (2019).
Crook, O. M., Mulvey, C. M., Kirk, P. D. W., Lilley, K. S. & Gatto, L. A Bayesian mixture modelling approach for spatial proteomics. PLoS Comput. Biol. 14, e1006516 (2018).
Lee, S. Y. et al. APEX fingerprinting reveals the subcellular localization of proteins of interest. Cell Rep. 15, 1837–1847 (2016).
Liu, X. et al. An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat. Commun. 9, 1188 (2018).
Go, C. D. et al. A proximity-dependent biotinylation map of a human cell. Nature 595, 120–124 (2021).
De Duve, C., Pressman, B. C., Gianetto, R., Wattiaux, R. & Appelmans, F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 60, 604–617 (1955).
Dunkley, T. P., Watson, R., Griffin, J. L., Dupree, P. & Lilley, K. S. Localization of organelle proteins by isotope tagging (LOPIT). Mol. Cell. Proteom. 3, 1128–1134 (2004).
Mulvey, C. M. et al. Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat. Protoc. 12, 1110–1135 (2017).
Liu, X. & Fagotto, F. A method to separate nuclear, cytosolic, and membrane-associated signaling molecules in cultured cells. Sci. Signal. 4, pl2 (2011).
Gatto, L., Breckels, L. M. & Lilley, K. S. Assessing sub-cellular resolution in spatial proteomics experiments. Curr. Opin. Chem. Biol. 48, 123–149 (2019).
Lund-Johansen, F. et al. MetaMass, a tool for meta-analysis of subcellular proteomics data. Nat. Methods 13, 837–840 (2016).
Binder, J. X. et al. COMPARTMENTS: unification and visualization of protein subcellular localization evidence. Database 2014, bau012 (2014).
Orsburn, B. C. Proteome Discoverer—a community enhanced data processing suite for protein informatics. Proteomes 9, 15 (2021).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301–2319 (2016).
Kong, A. T., Leprevost, F. V., Avtonomov, D. M., Mellacheruvu, D. & Nesvizhskii, A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017).
Holman, J. D., Tabb, D. L. & Mallick, P. Employing ProteoWizard to convert raw mass spectrometry data. Curr. Protoc. Bioinforma. 46, 13.24.1-9 (2014).
Kim, S. & Pevzner, P. A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277 (2014).
Granholm, V. et al. Fast and accurate database searches with MS-GF+Percolator. J. Proteome Res. 13, 890–897 (2014).
Sturm, M. et al. OpenMS – an open-source software framework for mass spectrometry. BMC Bioinforma. 9, 163 (2008).
Savitski, M. M., Wilhelm, M., Hahne, H., Kuster, B. & Bantscheff, M. A scalable approach for protein false discovery rate estimation in large proteomic data sets. Mol. Cell. Proteom. 14, 2394–2404 (2015).
Platt, J. Probabilistic outputs for support vector machines and comparisons to regularized likelihood methods. In Advances in Large Margin Classifiers (eds. Smola, A. J., Bartlett, P., Schölkopf, B. & Schuurmans, D.) (MIT Press, Cambridge, Massachusetts, USA, 1999).
Branca, R. M. et al. HiRIEF LC-MS enables deep proteome coverage and unbiased proteogenomics. Nat. Methods 11, 59–62 (2014).
Bantscheff, M. et al. Robust and sensitive iTRAQ quantification on an LTQ Orbitrap mass spectrometer. Mol. Cell. Proteom. 7, 1702–1713 (2008).
Ow, S. Y., Salim, M., Noirel, J., Evans, C. & Wright, P. C. Minimising iTRAQ ratio compression through understanding LC-MS elution dependence and high-resolution HILIC fractionation. Proteomics 11, 2341–2346 (2011).
Henderson, B. R. Nuclear-cytoplasmic shuttling of APC regulates β-catenin subcellular localization and turnover. Nat. Cell Biol. 2, 653–660 (2000).
Giurgiu, M. et al. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 47, D559–D563 (2019).
Bastian, M., Heymann, S. & Jacomy, M. Gephi: an open source software for exploring and manipulating networks. Proceedings of the Third International AAAI Conference on Weblogs and Social Media. (The AAAI Press, Menlo Park, California, USA, 2009).
Acknowledgements
This research was supported by funding from the Swedish Foundation for Strategic Research, Swedish Cancer Society, Swedish Research Council, Swedish Childhood Cancer Foundation, The Cancer Research Funds of Radiumhemmet and Stockholm’s County Council (ALF funding). We also thank O. Berkovska for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
The study was conceived by T.A., Y.P., M.V., L.M.O. and J.L. Cell culturing, subcellular fractionation and MS analysis were performed by Y.P. and G.M. Bioinformatics related to the classification of protein localization was performed by T.A. R package building was performed by T.A. Classification output analysis and evaluation of data in relation to other resources were performed by T.A. and L.M.O. The paper was written by T.A., Y.P., M.V. and L.M.O. All authors contributed in finalizing the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
J.L. reports receiving honoraria for invited speaker activities from Pfizer and Roche, institutional research support as a PI from AstraZeneca, Novartis and GE Healthcare. J.L. and L.M.O. are co-founders and shareholders of FenoMark Diagnostics AB.
Peer review
Peer review information
Nature Protocols thanks Josie Christopher, Markku Varjosalo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Orre, L. M. et al. Mol. Cell 73, 166–182.e7 (2019): https://doi.org/10.1016/j.molcel.2018.11.035
Key data used in this protocol
Orre, L. M. et al. Mol. Cell 73, 166–182.e7 (2019): https://doi.org/10.1016/j.molcel.2018.11.035
Extended data
Extended Data Fig. 1 Samples generated during cell fractionation.
a, Duplicate dishes of cells were incubated with digitonin (42 μg/ml) on a tilt rocker at 4 °C for 7 min. b, After incubation, the digitonin solution was recovered as fraction FS1. c, Exaggerated digitonin treatment (>10 min) will disrupt chromatin, resulting in DNA ‘threads’ in the dish upon harvesting of cells. d, Zoom in on DNA ‘threads’. e, After harvesting by scraping into low-salt buffer, the cells are transferred into a Dounce homogenizer. Mechanical disruption of cells is accomplished by repeated strokes into the homogenizer by a tight-fitting glass pestle. One stroke equals one down-and-up movement of the pestle. The strokes should be performed below the liquid level to avoid foaming. f, Examples of FP1 pellets generated by low-speed centrifugation. g, Examples of FP2 pellets generated by medium-speed centrifugation. For some cell lines, FP2 may be difficult to see by the naked eye. h, Examples of soluble fractions (FS2) and pellets (FP3) generated by ultracentrifugation at 100,000g for 1 h. sup., supernatant.
Extended Data Fig. 2 MS data comparison of MS approaches.
a, Cumulative distribution plots showing the number of PSMs used for protein quantification for different MS approaches. Indicated in the plots is the percentage of quantifications that were based on at least three PSMs. b, Scatter plots displaying the number of PSMs used for quantification and fractionation profile correlation between replicates. c, Bar plots showing the number of proteins identified (left, 1% FDR, gene centric), the number of unique peptides (middle, 1% FDR) and the number of PSMs used for quantification (right) for the three different MS approaches used. corr., correlation.
Extended Data Fig. 3 Classification output and MS method comparison.
a, Bar plots indicating the number of compartment classifications for the three different MS approaches used. b, Box plots displaying the classification probabilities for compartment-level classifications for the three different MS methods used. c, Box plots showing the minimum number of PSMs used for quantification of proteins on the basis of neighborhood classification agreement between HiRIEF and high-pH strategies (left) or HiRIEF and long-gradient strategies (right). Wilcoxon signed-rank test (two-sided) was used to calculate P values. d, Bar plots indicating the neighborhood classification agreement between methods for proteins quantified with one to two PSMs or more than two PSMs. e, Bar plots showing number of classifications and classification frequency of proteins binned by quantitative range (maximum value through minimum value in fractionation profile) for different MS approaches. Quantitative data are binned into five portions ranging between 0–0.5, 0.5–1, 1–1.5, 1.5–2 and >2. The elements of the boxplots in the figure are as follows: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers.
Extended Data Fig. 4 Comparison of classification output for different MS approaches.
a, Density plots showing the minimum number of PSMs used for quantifications of proteins. Density plots are colored on the basis of agreement between HiRIEF and high-pH MS strategies: yellow if protein classification is the same in both methods, red if it is different, blue if the protein is unclassified in one of the methods and green if unclassified in both methods. b, Scatter plot showing the maximum neighborhood classification probability score of HiRIEF and high-pH methods and the minimum number of PSMs used for quantification between HiRIEF and high-pH methods. Proteins are colored on the basis of classification agreement as described above (a). c, SubCellBarCodes for APC protein from the original five cell lines analyzed and available in the SubCellBarCode.org resource. The left figure indicates neighborhood-level classifications, and the right figure indicates compartment-level classifications. d, Scatter plots showing classification probability of proteins for HiRIEF and long-gradient MS strategies (left) and high-pH and long-gradient MS strategies (right). Proteins are colored on the basis of classification agreement: yellow indicates the same classification in both methods, red indicates a different classification, blue indicates proteins that were unclassified in one of the methods and green indicates proteins that were unclassified in both methods. class., classified; grad., gradient; Unclass., unclassified.
Supplementary information
Supplementary Information
Supplementary Method 1. BioConductor vignette for the SubCellBarCode R package
Supplementary Table 1
LC gradient lengths and strategy used for LC-MS analysis of the individual fractions (column A) generated by HiRIEF pre-fractionation in the pH ranges 3–10 (column B) and 3.4–4.8 (column C)
Supplementary Table 2
Relative quantitative data (TMT ratios) for the different MS datasets used in the current study. Data are represented in five different sheets: combined HiRIEF 3–10/3.4–4.8, HiRIEF 3–10, HiRIEF 3.4–4.8, high pH and long gradient. Each sheet includes the following columns: column A—gene symbol–centric protein ID, column B–L—TMT ratios for the five fractions (FS1, FS2 and FP1–3) in duplicate (A and B) and column M—minimum number of PSMs used for quantification for any of the 10 TMT channels
Supplementary Table 3
SubCellBarCode classification output for the different MS datasets used in the current study. Data are represented in five different sheets; combined HiRIEF 3–10/3.4–4.8, HiRIEF 3–10, HiRIEF 3.4–4.8, high pH and long gradient. Each sheet includes the following columns: column A—gene symbol–centric protein ID, column B—final neighborhood classification (SVMoutput), column C—final compartment classification (SVMoutput), columns D–G—SVM-derived probabilities for the individual neighborhoods and columns H–V—SVM-derived probabilities for the individual compartments
Source data
Source Data Fig. 4
Statistical and numerical source data
Source Data Fig. 5
Statistical and numerical source data
Source Data Fig. 6
Statistical and numerical source data
Source Data Fig. 7
statistical and numerical source data
Source Data Extended Data Fig. 2
Statistical and numerical source data
Source Data Extended Data Fig. 3
Statistical and numerical source data
Source Data Extended Data Fig. 4
Statistical and numerical source data
Rights and permissions
About this article
Cite this article
Arslan, T., Pan, Y., Mermelekas, G. et al. SubCellBarCode: integrated workflow for robust spatial proteomics by mass spectrometry. Nat Protoc 17, 1832–1867 (2022). https://doi.org/10.1038/s41596-022-00699-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00699-2
This article is cited by
-
Bioorthogonal photocatalytic proximity labeling in primary living samples
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.