Abstract
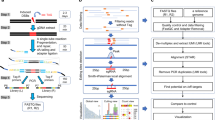
Genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq) is a sensitive, unbiased, genome-wide method for defining the activity of genome-editing nucleases in living cells. GUIDE-seq is based on the principle of efficient integration of an end-protected double-stranded oligodeoxynucleotide tag into sites of nuclease-induced DNA double-stranded breaks, followed by amplification of tag-containing genomic DNA molecules and high-throughput sequencing. Here we describe a detailed GUIDE-seq protocol including cell transfection, library preparation, sequencing and bioinformatic analysis. The entire protocol including cell culture can be completed in 9 d. Once tag-integrated genomic DNA is isolated, library preparation, sequencing and analysis can be performed in 3 d. The result is a genome-wide catalog of off-target sites ranked by nuclease activity as measured by GUIDE-seq read counts. GUIDE-seq is one of the most sensitive cell-based methods for defining genome-wide off-target activity and has been broadly adopted for research and therapeutic use.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data from experiments originally described in Lazzarotto et al.8 are available in NCBI Sequence Read Archive PRJNA625995.
Code availability
Open-source GUIDE-seq analysis code is available at https://github.com/tsailabSJ/guideseq.
References
Maeder, M. L. & Gersbach, C. A. Genome-editing technologies for gene and cell therapy. Mol. Ther. 24, 430–446 (2016).
Wang, H., Russa, M. L. & Qi, L. S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 85, 1–38 (2015).
Anderson, K. R. et al. CRISPR off-target analysis in genetically engineered rats and mice. Nat. Methods https://doi.org/10.1038/s41592-018-0011-5 (2018).
Tsai, S. Q. et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015).
Xie, K. & Yang, Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 6, 1975–1983 (2013).
Cheng, Y. & Tsai, S. Q. Illuminating the genome-wide activity of genome editors for safe and effective therapeutics. Genome Biol. 19, 226 (2018).
Putney, S., Benkovic, S. & Schimmel, P. A DNA fragment with an alpha-phosphorothioate nucleotide at one end is asymmetrically blocked from digestion by exonuclease III and can be replicated in vivo. Proc. Natl Acad. Sci. 78, 7350–7354 (1981).
Lazzarotto, C. R. et al. CHANGE-seq reveals genetic and epigenetic effects on CRISPR–Cas9 genome-wide activity. Nat. Biotechnol. 38, 1317–1327 (2020).
Zheng, Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 20, 1479–1484 (2014).
Tsai, S. Q., Topkar, V. V., Joung, K. J. & Aryee, M. J. Open-source guideseq software for analysis of GUIDE-seq data. Nat. Biotechnol. 34, 483–483 (2016).
Tsai, S. Q. et al. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614 (2017).
Wienert, B. et al. Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289 (2019).
Wang, X. et al. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 33, 175–178 (2015).
Kim, D. et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 12, 237–243 (2015).
Yan, W. X. et al. BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun. 8, 15058 (2017).
Schmid-Burgk, J. L. et al. Highly parallel profiling of Cas9 variant specificity. Mol. Cell 78, 794–800.e8 (2020).
Crosetto, N. et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365 (2013).
Wang, L. et al. Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat. Biotechnol. 36, 717–725 (2018).
Miller, J. C. et al. Enhancing gene editing specificity by attenuating DNA cleavage kinetics. Nat. Biotechnol. 37, 945–952 (2019).
Kleinstiver, B. et al. Genome-wide specificities of CRISPR-Cas Cpf1 nucleases in human cells. Nat. Biotechnol. 34, 869.
Nobles, C. L. et al. iGUIDE: an improved pipeline for analyzing CRISPR cleavage specificity. Genome Biol. 20, 14 (2019).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science https://doi.org/10.1126/science.aba8853 (2020).
Hendel, A. et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 33, 985–989 (2015).
Fiume, M. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019).
Acknowledgements
This work was supported by St. Jude Children’s Research Hospital and ALSAC, National Institutes of Health (NIH) Office Of The Director (OD) Somatic Cell Genome Editing (SCGE) initiative grant U01AI157189 (to S.Q.T.), St. Jude Children’s Research Hospital Collaborative Research Consortium on Novel Gene Therapies for Sickle Cell Disease (SCD), the Doris Duke Charitable Foundation (2017093 and 2020154) (to S.Q.T.), an (NIH) Directors Pioneer Award (DP1 GM105378) (to J.K.J.), NIH R01 GM088040 (to J.K.J.), NIH R01 AR063070 (to J.K.J.), and the Jim and Ann Orr Massachusetts General Hospital (MGH) Research Scholar Award (to J.K.J.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Contributions
N.L.M., C.R.L., G.L., Y.L. and S.Q.T. drafted the manuscript. S.Q.T. and J.K.J. conceived the GUIDE-seq method. S.Q.T. and Z.Z. developed the GUIDE-seq protocol in the laboratories of J.K.J. and A.I. M.L. and N.N. performed experiments during original development of GUIDE-seq under supervision of S.Q.T. and Z.Z. S.Q.T., V.T. and M.A. contributed to development of the GUIDE-seq analysis software.
Corresponding author
Ethics declarations
Competing interests
S.Q.T. is a co-inventor on patents covering the GUIDE-seq method. S.Q.T. is a member of the scientific advisory board of Kromatid and Twelve Bio. M.J.A. and J.K.J. hold equity in SeQure Dx, Inc. J.K.J. holds equity in Chroma Medicine. J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Excelsior Genomics, Pairwise Plants, Poseida Therapeutics, Transposagen Biopharmaceuticals, and Verve Therapeutics (f/k/a Endcadia). J.K.J.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. is a co-inventor on various patents and patent applications that describe gene editing and epigenetic editing technologies, including the GUIDE-seq method.
Additional information
Peer review information Nature Protocols thanks Ciaran M. Lee and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Tsai, S. Q. et al. Nat. Biotechnol. 33, 187 (2015): https://doi.org/10.1038/nbt.3117
Lazzarotto, C. R. et al. Nat. Biotechnol. 38, 1317 (2020): https://doi.org/10.1038/s41587-020-0555-7
Kleinstiver P. B. et al. Nat. Biotechnol. 34 869 (2016): https://doi.org/10.1038/nbt.3620
Supplementary information
Supplementary Data
Example Illumina SampleSheet SampleSheet for NGS sequencing of GUIDE-seq samples. Demultiplexing is built into the pipeline and library’s indices shouldn’t be specified here. All sequence reads will be sorted into an Undetermined category used for further processing. However, it is important to add an extra eight As after index 2 to include reading of UMI required by the pipeline.
Rights and permissions
About this article
Cite this article
Malinin, N.L., Lee, G., Lazzarotto, C.R. et al. Defining genome-wide CRISPR–Cas genome-editing nuclease activity with GUIDE-seq. Nat Protoc 16, 5592–5615 (2021). https://doi.org/10.1038/s41596-021-00626-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00626-x
This article is cited by
-
Whole-brain in vivo base editing reverses behavioral changes in Mef2c-mutant mice
Nature Neuroscience (2024)
-
An engineered hypercompact CRISPR-Cas12f system with boosted gene-editing activity
Nature Chemical Biology (2023)
-
A cleavage rule for selection of increased-fidelity SpCas9 variants with high efficiency and no detectable off-targets
Nature Communications (2023)
-
Versatile and efficient genome editing with Neisseria cinerea Cas9
Communications Biology (2022)
-
TAPE-seq is a cell-based method for predicting genome-wide off-target effects of prime editor
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.