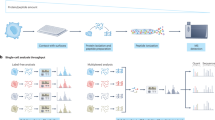
Abstract
Many biological systems are composed of diverse single cells. This diversity necessitates functional and molecular single-cell analysis. Single-cell protein analysis has long relied on affinity reagents, but emerging mass-spectrometry methods (either label-free or multiplexed) have enabled quantifying >1,000 proteins per cell while simultaneously increasing the specificity of protein quantification. Here we describe the Single Cell ProtEomics (SCoPE2) protocol, which uses an isobaric carrier to enhance peptide sequence identification. Single cells are isolated by FACS or CellenONE into multiwell plates and lysed by Minimal ProteOmic sample Preparation (mPOP), and their peptides labeled by isobaric mass tags (TMT or TMTpro) for multiplexed analysis. SCoPE2 affords a cost-effective single-cell protein quantification that can be fully automated using widely available equipment and scaled to thousands of single cells. SCoPE2 uses inexpensive reagents and is applicable to any sample that can be processed to a single-cell suspension. The SCoPE2 workflow allows analyzing ~200 single cells per 24 h using only standard commercial equipment. We emphasize experimental steps and benchmarks required for achieving quantitative protein analysis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data are available at MassIVE MSV000087041 and via the SCoPE2 website: https://scope2.slavovlab.net/mass-spec/protocol. Datasets associated with certain figures have been uploaded to figshare: Fig. 3: Slavov, Nikolai (2021): Evaluating data acquisition and interpretation using diagnostic plots generated by DO-MS (https://doi.org/10.6084/m9.figshare.15060774.v1); Fig. 4: Slavov, Nikolai (2021): Principal Component Analysis (PCA) of quantification results from SCoPE2 analysis (https://doi.org/10.6084/m9.figshare.15060789.v1); Extended Data Fig. 1: Slavov, Nikolai (2021): The ABIRD device may suppress contaminant ions and enhance peptide identification (https://doi.org/10.6084/m9.figshare.15060846.v1) and Slavov, Nikolai (2021): Variable TMT search indicates very low level of over labeled peptides during SCoPE2 analysis (https://doi.org/10.6084/m9.figshare.15145092.v1). Source data are provided with this paper.
Code availability
The SCoPE2 pipeline used here is available at https://github.com/SlavovLab/SCoPE2 and via the SCoPE2 website (https://scope2.slavovlab.net/mass-spec/protocol).
References
Symmons, O. & Raj, A. What’s luck got to do with it: single cells, multiple fates, and biological nondeterminism. Mol. Cell 62, 788–802 (2016).
Paul, I., White, C., Turcinovic, I. & Emili, A. Imaging the future: the emerging era of single-cell spatial proteomics. FEBS J. 2020).
Levy, E. & Slavov, N. Single cell protein analysis for systems biology. Essays Biochem. https://doi.org/10.1042/EBC20180014 (2018).
Regev, A. et al. Science forum: the human cell atlas. eLife 6, e27041 (2017).
Ziegenhain, C. et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643 (2017).
Engblom, C. et al. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science 358 2017).
Shaffer, S. M. et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature 546, 431–435 (2017).
Franks, A., Airoldi, E. & Slavov, N. Post-transcriptional regulation across human tissues. PLoS Comput. Biol. 13, e1005535 (2017).
Slavov, N. Unpicking the proteome in single cells. Science 367, 512–513 (2020).
Washburn, M. P., Wolters, D. & Yates, J. R. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 (2001).
Altelaar, A. F. M., Munoz, J. & Heck, A. J. R. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat. Rev. Genet. 14, 35–48 (2013).
Cravatt, B. F., Simon, G. M. & Yates Iii, J. R. The biological impact of mass-spectrometry-based proteomics. Nature 450, 991 (2007).
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A. J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Muntel, J. et al. Comparison of protein quantification in a complex background by DIA and TMT workflows with fixed instrument time. J. Proteome Res. 18, 1340–1351 (2019). PMID: 30726097.
Slavov, N. Single-cell protein analysis by mass spectrometry. Curr. Opin. Chem. Biol. 60, 1–9 (2020).
Kelly, R. T. Single-cell proteomics: progress and prospects. Mol. Cell. Proteomics 19, 1739–1748 (2020).
Budnik, B., Levy, E., Harmange, G. & Slavov, N. SCoPE-MS: mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biology 19, 161 (2018).
Specht, H. et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol. https://doi.org/10.1186/s13059-021-02267-5 (2021).
Specht, H. & Slavov, N. Optimizing accuracy and depth of protein quantification in experiments using isobaric carriers. J. Proteome Res. 20, a8802021–887 (2021).
Cheung, T. K. et al. Defining the carrier proteome limit for single-cell proteomics. Nat. Methods 18, 76–83 (2021).
Specht, H. et al. Automated sample preparation for high-throughput single-cell proteomics. Preprint at bioRxiv https://doi.org/10.1101/399774 (2018).
Marx, V. A dream of single-cell proteomics. Nat. Methods 16, 809–812 (2019).
Leduc, A., Huffman, R. G. & Slavov, N. Droplet sample preparation for single-cell proteomics applied to the cell cycle. Preprint at bioRxiv https://doi.org/10.1101/2021.04.24.441211 (2021).
Chen, A. T., Franks, A. & Slavov, N. DART-ID increases single-cell proteome coverage. PLOS Comput. Biol. 15, 1–30 (2019).
Huffman, G., Chen, A. T., Specht, H. & Slavov, N. DO-MS: data-driven optimization of mass spectrometry methods. J. Proteome Res. https://doi.org/10.1021/acs.jproteome.9b00039 (2019).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Albayrak, C. et al. Digital quantification of proteins and mRNA in single mammalian cells. Mol. Cell 61, 914–924 (2016).
Darmanis, S. et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 14, 380–389 (2016).
Hughes, A. J. et al. Single-cell western blotting. Nat. Methods 11, 749–755 (2014).
Specht, H., Emmott, E., Perlman, D. H., Koller, A. & Slavov, N. High-throughput single-cell proteomics quantifies the emergence of macrophage heterogeneity. Preprint at bioRxiv. https://doi.org/10.1101/665307 (2019).
Virant-Klun, I., Leicht, S., Hughes, C. & Krijgsveld, J. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol. Cell. Proteomics 15, 2616–2627 (2016).
Lombard-Banek, C., Moody, S. A. & Nemes, P. Single-cell mass spectrometry for discovery proteomics: quantifying translational cell heterogeneity in the 16-cell frog (Xenopus) embryo. Angew. Chem. Int. Ed. 55, 2454–2458 (2016).
Cong, Y. et al. Improved single-cell proteome coverage using narrow-bore packed nanoLC columns and ultrasensitive mass spectrometry. Anal. Che. 92, 2665–2671 (2020).
Cong, Y. et al. Ultrasensitive single-cell proteomics workflow identifies >1000 protein groups per mammalian cell. Chem. Sci. 12, 1001–1006 (2020).
Brunner, A.-D. et al. Ultra-high sensitivity mass spectrometry quantifies single-cell proteome changes upon perturbation. Preprint at bioRxiv https://doi.org/10.1101/2020.12.22.423933 (2020).
Zhu, Y. et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun. 9, 882 (2018).
Dou, M. et al. High-throughput single cell proteomics enabled by multiplex isobaric labeling in a nanodroplet sample preparation platform. Anal. Chem. 91, 13119–13127 (2019).
Li, Z.-Y. et al. Nanoliter-scale oil-air-droplet chip-based single cell proteomic analysis. Anal. Chem. 90, 5430–5438 (2018).
Liang, Y. et al. Fully automated sample processing and analysis workflow for low-input proteome profiling. Anal. Chem. 93, 1658–1666 (2021).
Lafzi, A., Moutinho, C., Picelli, S. & Heyn, H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 13, 2742–2757 (2018).
Barkas, N. et al. Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat. Methods 16, 695–698 (2019).
Marx, V. A dream of single-cell proteomics. Nat. Methods. https://doi.org/10.1038/s41592-019-0540-6 (2019).
Grün, D. & van Oudenaarden, A. Design and analysis of single-cell sequencing experi- ments. Cell 163, 799–810 (2015).
Meier, F. et al. Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545 (2018).
Specht, H. & Slavov, N. Transformative opportunities for single-cell proteomics. J. Proteome Res. 17, 2563–2916 (2018).
Slavov, N. Driving single cell proteomics forward with innovation. J. Proteome Res. https://doi.org/10.1021/acs.jproteome.1c00639 (2021).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized ppb- range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11, 2301 (2016).
Yu, S.-H., Kyriakidou, P. & Cox, J. Isobaric matching between runs and novel PSM-level normalization in MaxQuant strongly improve reporter ion-based quantification. J. Proteome Res. 19, 3945–3954 (2020).
Fondrie, W. E. & Noble, W.S. mokapot: fast and flexible semisupervised learning for peptide detection. J. Proteome Res. 20, 1966–1971 (2021).
Milo, R., Jorgensen, P., Moran, U., Weber, G. & Springer, M. BioNumbers—the database of key numbers in molecular and cell biology. Nucleic Acids Res. 38, D750–D753 (2010).
Specht, H. et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity. GitHub github.com/SlavovLab/SCoPE2 (2019).
Specht, H. et al. Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Zenodo https://doi.org/10.5281/zenodo.4339954 (2020).
Vanderaa, C. & Gatto, L. Replication of single-cell proteomics data reveals important computational challenges. Preprint at bioRxiv. https://doi.org/10.1101/2021.04.12.439408 (2021).
Vanderaa, C. & Gatto, L. Mass spectrometry-based single-cell proteomics data analysis. Bioconductor http://www.bioconductor.org/packages/release/bioc/html/scp.html (2020).
SCP2019 Workshop on single-cell proteomics Aug. 2019. http://workshop2019.single-cell.net
Acknowledgements
We thank A. Chen, and J. Neveu for assistance, discussions and constructive comments. This work was funded by a New Innovator Award from the NIGMS from the National Institutes of Health to N.S. under Award Number DP2GM123497, an Allen Distinguished Investigator award through The Paul G. Allen Frontiers Group to N.S., a Seed Networks Award from CZI CZF2019-002424 to N.S., through a Merck Exploratory Science Center Fellowship, Merck Sharpe & Dohme Corp. to N.S., and a Thermo Scientific Tandem Mass Tag Research Award to E.E. Funding bodies had no role in data collection, analysis and interpretation.
Author information
Authors and Affiliations
Contributions
Experimental design: A.P. and N.S. Sorting cells: A.L. and A.P. LC-MS/MS: G.H, E.E., H.S. and D.H.P. Sample preparation: A.P. Funding: N.S, E.E. Data analysis: A.P. and N.S. Supervision: N.S. Writing and editing: E.E., A.P. and N.S.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks M. Arthur Moseley, Wenqing Shui and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Specht, H. et al. Genome Biol. 22, 50 (2021): https://doi.org/10.1186/s13059-021-02267-5
Budnik, B. et al. Genome Biol. 19, 161 (2018): https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1547-5
Specht, H. & Slavov, N. J. Proteome Res. 20, 880887 (2021): https://pubs.acs.org/doi/10.1021/acs.jproteome.0c00675
Extended data
Extended Data Fig. 1 The ABIRD may suppress contaminant ions and enhance peptide identification.
ABIRD may suppress contaminant ions and enhance peptide identification. Replicate injections of a 1× standard were analyzed with the ABIRD on or off. a, The replicates with ABIRD on had a reduced number of +1 ions (likely corresponding to contaminants) and an increased number of higher-charge-state ions, which are likely to correspond to peptides. b, With the ABIRD on, the number of identified peptides is increased across all confidence levels (PEP).
Supplementary information
Supplementary Data 1
XML file for MaxQuant that specifies the parameters for TMTpro 16-plex searches.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Rights and permissions
About this article
Cite this article
Petelski, A.A., Emmott, E., Leduc, A. et al. Multiplexed single-cell proteomics using SCoPE2. Nat Protoc 16, 5398–5425 (2021). https://doi.org/10.1038/s41596-021-00616-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00616-z
This article is cited by
-
The 37TrillionCells initiative for improving global healthcare via cell-based interception and precision medicine: focus on neurodegenerative diseases
Molecular Brain (2024)
-
Pick-up single-cell proteomic analysis for quantifying up to 3000 proteins in a Mammalian cell
Nature Communications (2024)
-
scPROTEIN: a versatile deep graph contrastive learning framework for single-cell proteomics embedding
Nature Methods (2024)
-
A critical evaluation of ultrasensitive single-cell proteomics strategies
Analytical and Bioanalytical Chemistry (2024)
-
Sampling the proteome by emerging single-molecule and mass spectrometry methods
Nature Methods (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.