Abstract
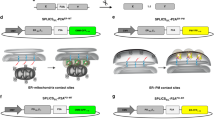
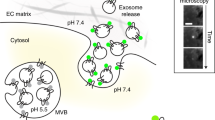
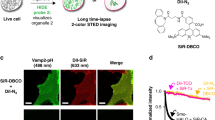
Membrane contact sites between organelles are essential for maintaining cellular homeostasis, which requires the continuous exchange of signaling molecules, ions, nutrients and lipids. Alterations of different contact sites are associated with a wide spectrum of human diseases. However, visualizing and quantifying these contact sites remains a challenge. This protocol describes the use of split-GFP-based contact site sensors (SPLICS) in microscopy applications for mapping organelle contact sites both in fixed and living cells. SPLICS sensors are engineered to express equimolar amounts of the organelle-targeted nonfluorescent β11 and GFP1-10 portions of the split-GFP protein in a single vector, and are capable of reconstituting fluorescence when two opposing membranes come into proximity. Reconstitution will occur only over the cell volume at defined contact sites resulting in a bright signal that can be detected easily and quantified automatically with specific custom-made plugins. The use of minimal targeting sequences facilitates targeting specificity and membrane coverage, avoiding artifacts due to full-length fusion protein overexpression and, thus, possible perturbations of the cell’s physiology. SPLICS sensors engineered to simultaneously detect multiple contact sites within the same cell have been generated by exploiting the ability of the β11 GFP fragment to reconstitute different color-shifted variants of the GFP1-10 fragment. Here, we describe a detailed protocol to set up SPLICS expression in living cells (2–3 d), detection and acquisition (1 d), and automated quantification with custom plugins (1–2 d). We also advise on construct design and characterization for novel organelle contacts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Code availability
Quantification plugins are available at https://github.com/titocali1/Quantification-Plugins47.
References
Berenguer-Escuder, C. et al. Variants in Miro1 cause alterations of ER-mitochondria contact sites in fibroblasts from Parkinson’s disease patients. J. Clin. Med. https://doi.org/10.3390/jcm8122226 (2019).
Cali, T. et al. splitGFP technology reveals dose-dependent ER-mitochondria interface modulation by α-synuclein A53T and A30P mutants. Cells 8, 1072 (2019).
Carreras-Sureda, A. et al. Non-canonical function of IRE1alpha determines mitochondria-associated endoplasmic reticulum composition to control calcium transfer and bioenergetics. Nat. Cell Biol. 21, 755–767 (2019).
Cieri, D. et al. SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ. 25, 1131–1145 (2018).
Doghman-Bouguerra, M. et al. FATE1 antagonizes calcium- and drug-induced apoptosis by uncoupling ER and mitochondria. EMBO Rep. 17, 1264–1280 (2016).
Filadi, R. et al. TOM70 sustains cell bioenergetics by promoting IP3R3-mediated ER to mitochondria Ca2+ transfer. Curr. Biol. 28, 369–382 e366 (2018).
Gomez-Suaga, P. et al. The VAPB-PTPIP51 endoplasmic reticulum-mitochondria tethering proteins are present in neuronal synapses and regulate synaptic activity. Acta Neuropathol. Commun. 7, 35 (2019).
Granatiero, V. et al. Reduced mitochondrial Ca2+ transients stimulate autophagy in human fibroblasts carrying the 13514A>G mutation of the ND5 subunit of NADH dehydrogenase. Cell Death Differ. 23, 231–241 (2016).
Gutierrez, T. et al. The ER chaperone calnexin controls mitochondrial positioning and respiration. Sci. Signal. https://doi.org/10.1126/scisignal.aax6660 (2020).
Hewitt, V. L. et al. Decreasing pdzd8-mediated mitochondrial-ER contacts in neurons improves fitness by increasing mitophagy. Preprint at bioRxiv https://doi.org/10.1101/2020.11.14.382861 (2020).
Vallese, F. et al. An expanded palette of improved SPLICS reporters detects multiple organelle contacts in vitro and in vivo. Nat. Commun. 11, 6069 (2020).
Yeshaw, W. M. et al. Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. eLife 8, https://doi.org/10.7554/eLife.43561 (2019).
Cieri, D. et al. Tau localises within mitochondrial sub-compartments and its caspase cleavage affects ER-mitochondria interactions and cellular Ca2+ handling. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 3247–3256 (2018).
Dematteis, G. et al. Proteomic analysis links alterations of bioenergetics, mitochondria-ER interactions and proteostasis in hippocampal astrocytes from 3xTg-AD mice. Cell Death Dis. 11, 645 (2020).
Naia, L. et al. Neuronal cell-based high-throughput screen for enhancers of mitochondrial function reveals luteolin as a modulator of mitochondria-endoplasmic reticulum coupling. BMC Biol. https://doi.org/10.1186/s12915-021-00979-5 (2021).
Ciscato, F. et al. Hexokinase 2 displacement from mitochondria-associated membranes prompts Ca2+-dependent death of cancer cells. EMBO Rep. 21, e49117 (2020).
Lopez-Crisosto, C. et al. Endoplasmic reticulum–mitochondria coupling increases during doxycycline-induced mitochondrial stress in HeLa cells. Cell Death Dis. 12, 657 (2021).
Cabantous, S., Terwilliger, T. C. & Waldo, G. S. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 (2005).
Pedelacq, J. D., Cabantous, S., Tran, T., Terwilliger, T. C. & Waldo, G. S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88 (2006).
Cabantous, S. & Waldo, G. S. In vivo and in vitro protein solubility assays using split GFP. Nat. Methods 3, 845–854 (2006).
Ryan, M. D., King, A. M. & Thomas, G. P. Cleavage of foot-and-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. J. Gen. Virol. 72, 2727–2732 (1991).
Csordas, G. et al. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915–921 (2006).
Giacomello, M. et al. Ca2+ hot spots on the mitochondrial surface are generated by Ca2+ mobilization from stores, but not by activation of store-operated Ca2+ channels. Mol. Cell 38, 280–290 (2010).
Gonzalo, S., Greentree, W. K. & Linder, M. E. SNAP-25 is targeted to the plasma membrane through a novel membrane-binding domain. J. Biol. Chem. 274, 21313–21318 (1999).
Macpherson, L. J. et al. Dynamic labelling of neural connections in multiple colours by trans-synaptic fluorescence complementation. Nat. Commun. 6, 10024 (2015).
Silva, B. S. C. et al. Maintaining social contacts: the physiological relevance of organelle interactions. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118800 (2020).
Filadi, R. et al. Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proc. Natl Acad. Sci. USA 112, E2174–E2181 (2015).
Rizzuto, R., Brini, M., Murgia, M. & Pozzan, T. Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science 262, 744–747 (1993).
Brunstein, M., Wicker, K., Herault, K., Heintzmann, R. & Oheim, M. Full-field dual-color 100-nm super-resolution imaging reveals organization and dynamics of mitochondrial and ER networks. Opt. Express 21, 26162–26173 (2013).
Bottanelli, F. et al. Two-colour live-cell nanoscale imaging of intracellular targets. Nat. Commun. 7, 10778 (2016).
Alford, S. C., Abdelfattah, A. S., Ding, Y. & Campbell, R. E. A fluorogenic red fluorescent protein heterodimer. Chem. Biol. 19, 353–360 (2012).
Alford, S. C., Ding, Y., Simmen, T. & Campbell, R. E. Dimerization-dependent green and yellow fluorescent proteins. ACS Synth. Biol. 1, 569–575 (2012).
Tchekanda, E., Sivanesan, D. & Michnick, S. W. An infrared reporter to detect spatiotemporal dynamics of protein-protein interactions. Nat. Methods 11, 641–644 (2014).
Csordas, G. et al. Imaging interorganelle contacts and local calcium dynamics at the ER-mitochondrial interface. Mol. Cell 39, 121–132 (2010).
Kakimoto, Y. et al. Visualizing multiple inter-organelle contact sites using the organelle-targeted split-GFP system. Sci. Rep. 8, 6175 (2018).
Copeland, D. E. & Dalton, A. J. An association between mitochondria and the endoplasmic reticulum in cells of the pseudobranch gland of a teleost. J. Biophys. Biochem. Cytol. 5, 393–396 (1959).
Bernhard, W. & Rouiller, C. Close topographical relationship between mitochondria and ergastoplasm of liver cells in a definite phase of cellular activity. J. Biophys. Biochem. Cytol. 2, 73–78 (1956).
Wieckowski, M. R., Giorgi, C., Lebiedzinska, M., Duszynski, J. & Pinton, P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat. Protoc. 4, 1582–1590 (2009).
Cho, I. T. et al. Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum-mitochondrial contacts. J. Biol. Chem. 292, 16382–16392 (2017).
Hung, V. et al. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife https://doi.org/10.7554/eLife.24463 (2017).
Soderberg, O. et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 (2006).
Ravikumar, B., Duden, R. & Rubinsztein, D. C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 11, 1107–1117 (2002).
Shi, F. et al. Optogenetic control of endoplasmic reticulum-mitochondria tethering. ACS Synth. Biol. 7, 2–9 (2018).
Yang, Z., Zhao, X., Xu, J., Shang, W. & Tong, C. A novel fluorescent reporter detects plastic remodeling of mitochondria-ER contact sites. J. Cell Sci. https://doi.org/10.1242/jcs.208686 (2018).
Harmon, M., Larkman, P., Hardingham, G., Jackson, M. & Skehel, P. A bi-fluorescence complementation system to detect associations between the endoplasmic reticulum and mitochondria. Sci. Rep. 7, 17467 (2017).
Shai, N. et al. Systematic mapping of contact sites reveals tethers and a function for the peroxisome-mitochondria contact. Nat. Commun. 9, 1761 (2018).
Cali, T. & Brini, M. Quantification of organelle contact sites by split-GFP-based contact site sensors (SPLICS) in living cells. Zenodo https://doi.org/10.5281/zenodo.5031309 (2021).
Ottolini, D., Cali, T. & Brini, M. Methods to measure intracellular Ca2+ fluxes with organelle-targeted aequorin-based probes. Methods Enzymol. 543, 21–45 (2014).
Ottolini, D., Cali, T. & Brini, M. Measurements of Ca2+ concentration with recombinant targeted luminescent probes. Methods Mol. Biol. 937, 273–291 (2013).
Graham, F. L. & van der Eb, A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52, 456–467 (1973).
Acknowledgements
The work is supported by grants from the Ministry of University and Research (Bando SIR 2014 no. RBSI14C65Z and PRIN2017 to T.C.) and from the Università degli Studi di Padova (Progetto Giovani 2012 no. GRIC128SP0 to T.C., Progetto di Ateneo 2016 no. CALI_SID16_01 to T.C., STARS Consolidator Grant 2019 to T.C. and Progetto di Ateneo 2015 no. CPDA153402 to M.B.).
Author information
Authors and Affiliations
Contributions
T.C. and M.B. equally contributed to the conceptualization, data curation, methodology, writing-original draft preparation, revising and editing and funding acquisition of the present manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Stéphanie Cabantous and the other, anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Cieri, D. et al. Cell Death Differ. 25, 1131–1145 (2018): https://doi.org/10.1038/s41418-017-0033-z
Vallese, F. et al. Nat. Commun. 11, 6069 (2020): https://doi.org/10.1038/s41467-020-19892-6
Cali, T. et al. Cells 8, 1072 (2019): https://doi.org/10.3390/cells8091072
Supplementary information
Rights and permissions
About this article
Cite this article
Calì, T., Brini, M. Quantification of organelle contact sites by split-GFP-based contact site sensors (SPLICS) in living cells. Nat Protoc 16, 5287–5308 (2021). https://doi.org/10.1038/s41596-021-00614-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00614-1
This article is cited by
-
A SPLICS reporter reveals \({{{{{\boldsymbol{\alpha }}}}}}\)-synuclein regulation of lysosome-mitochondria contacts which affects TFEB nuclear translocation
Nature Communications (2024)
-
Perturbation of the host cell Ca2+ homeostasis and ER-mitochondria contact sites by the SARS-CoV-2 structural proteins E and M
Cell Death & Disease (2023)
-
Integrated multimodality microscope for accurate and efficient target-guided cryo-lamellae preparation
Nature Methods (2023)
-
Protein synthesis inhibition and loss of homeostatic functions in astrocytes from an Alzheimer’s disease mouse model: a role for ER-mitochondria interaction
Cell Death & Disease (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.