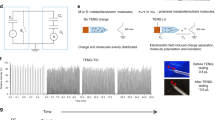
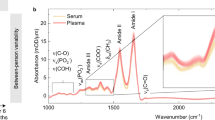
Abstract
Of the many metabolites involved in any clinical condition, only a narrow range of biomarkers is currently being used in the clinical setting. A key to personalized medicine would be to extend this range. Metabolic fingerprinting provides a more comprehensive insight, but many methods used for metabolomics analysis are too complex and time-consuming to be diagnostically useful. Here, a rapid evaporative ionization mass spectrometry (REIMS) system for direct ex vivo real-time analysis of biofluids with minor sample pretreatment is detailed. The REIMS can be linked to various laser wavelength systems (such as optical parametric oscillator or CO2 laser) and with automation for high-throughput analysis. Laser-induced sample evaporation occurs within seconds through radiative heating with the plume guided to the MS instrument. The presented procedure includes (i) laser setup with automation, (ii) analysis of biofluids (blood/urine/stool/saliva/sputum/breast milk) and (iii) data analysis. We provide the optimal settings for biofluid analysis and quality control, enabling sensitive, precise and robust analysis. Using the automated setup, 96 samples can be analyzed in ~35–40 min per ionization mode, with no intervention required. Metabolic fingerprints are made up of 2,000–4,000 features, for which relative quantification can be achieved at high repeatability when total ion current normalization is applied. With saliva and feces as example matrices, >70% of features had a coefficient of variance ≤30%. However, to achieve acceptable long-term reproducibility, additional normalizations by, e.g., LOESS are recommended, especially for positive ionization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Datasets relevant to our published supporting primary papers can be made available from the corresponding author upon reasonable request. The source data for figures are publicly available in the Figshare repository: Fig. 3, https://doi.org/10.6084/m9.figshare.14258423.v1; Fig. 5, https://doi.org/10.6084/m9.figshare.14258438.v1; Fig. 6 https://doi.org/10.6084/m9.figshare.14258459.v2.
References
Gong, Z., Zhang, J. & Xu, Y.-J. Metabolomics reveals that Momordica charantia attenuates metabolic changes in experimental obesity. Phyther. Res 31, 296–302 (2017).
Pham-Tuan, H., Kaskavelis, L., Daykin, C. A. & Janssen, H. G. Method development in high-performance liquid chromatography for high-throughput profiling and metabonomic studies of biofluid samples. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 789, 283–301 (2003).
Wishart, D. S. Metabolomics for investigating physiological and pathophysiological processes. Physiol. Rev. 99, 1819–1875 (2019).
Johnson, C. H., Ivanisevic, J. & Siuzdak, G. Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17, 451–459 (2016).
Ferreira, C. R. et al. Ambient ionization mass spectrometry for point-of-care diagnostics and other clinical measurements. Clin. Chem. 62, 99–110 (2016).
Cameron, S. J. S. et al. Evaluation of direct from sample metabolomics of human feces using rapid evaporative ionization mass spectrometry. Anal. Chem. 91, 13448–13457 (2019).
Ogrinc, N. et al. Water-assisted laser desorption/ionization mass spectrometry for minimally invasive in vivo and real-time surface analysis using SpiderMass. Nat. Protoc. 14, 3162–3182 (2019).
Schäfer, K. C. et al. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry. Angew. Chem. Int. Ed. 48, 8240–8242 (2009).
Clendinen, C. S., Monge, M. E. & Fernández, F. M. Ambient mass spectrometry in metabolomics. Analyst 142, 3101–3117 (2017).
Li, L., Hsieh, H. & Hsu, C. Clinical application of ambient ionization. Mass Spectrom. (Tokyo) 6, Spec Iss S0060 (2017).
Van Meulebroek, L. et al. Rapid LA-REIMS and comprehensive UHPLC-HRMS for metabolic phenotyping of feces. Talanta 217, 121043 (2020).
Wijnant, K. et al. Validated ultra-high-performance liquid chromatography hybrid high- resolution mass spectrometry and laser-assisted rapid evaporative ionization mass spectrometry for salivary metabolomics. Anal. Chem. 92, 5116–5124 (2020).
Fatou, B. et al. In vivo real-time mass spectrometry for guided surgery application. Sci. Rep. 6, 25919 (2016).
Kuo, T. H., Dutkiewicz, E. P., Pei, J. & Hsu, C. C. Ambient ionization mass spectrometry today and tomorrow: embracing challenges and opportunities. Anal. Chem. 92, 2353–2363 (2020).
Schäfer, K.-C. et al. In situ, real-time identification of biological tissues by ultraviolet and infrared laser desorption ionization mass spectrometry. Anal. Chem. 83, 1632–1640 (2011).
Balog, J. et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med. 5, 1–11 (2013).
Feider, C. L., Krieger, A., Dehoog, R. J. & Eberlin, L. S. Ambient ionization mass spectrometry: recent developments and applications. Anal. Chem. 91, 4266–4290 (2019).
Fillet, M. & Fre, M. Metabolomics in medicinal chemistry. The emergence of metabolomics as a key discipline in the drug discovery process. Drug Discov. Today Technol. 13, 19–24 (2015).
Golf, O. et al. Rapid evaporative ionization mass spectrometry imaging platform for direct mapping from bulk tissue and bacterial growth media. Anal. Chem. 87, 2527–2534 (2015).
Nemes, P. & Vertes, A. Ambient mass spectrometry for in vivo local analysis and in situ molecular tissue imaging. Trends Analyt. Chem. 34, 22–34 (2012).
Jones, E. A. et al. Matrix assisted rapid evaporative ionization mass spectrometry. Anal. Chem. 91, 9784–9791 (2019).
Suva, M. A brief review on dried blood spots applications in drug development. J. Pharm. Biosci. 2, 17–23 (2014).
Van Meulebroek, L. et al. Holistic lipidomics of the human gut phenotype using validated ultra-high-performance liquid chromatography coupled to hybrid orbitrap mass spectrometry. Anal. Chem. 22, 12502–12510 (2017).
Zhao, J., Evans, C. R., Carmody, L. A. & LiPuma, J. J. Impact of storage conditions on metabolite profiles of sputum samples from persons with cystic fibriosis. J. Cyst. Fibros. 14, 468–473 (2015).
De Paepe, E. et al. A validated multi-matrix platform for metabolomic fingerprinting of human urine, feces and plasma using ultra-high performance liquid-chromatography coupled to hybrid orbitrap high-resolution mass spectrometry. Anal. Chim. Acta 1033, 108–118 (2018).
Cameron, S. J. S. & Takáts, Z. Mass spectrometry approaches to metabolic profiling of microbial communities within the human gastrointestinal tract. Methods (San. Diego, Calif.) 149, 13–24 (2018).
Gowers, G. O. F. et al. Off-colony screening of biosynthetic libraries by rapid laser-enabled mass spectrometry. ACS Synth. Biol. 8, 2566–2575 (2019).
Williams, J. P. & Scrivens, J. H. Coupling desorption electrospray ionisation and neutral desorption/extractive electrospray ionisation with a travelling-wave based ion mobility mass spectrometer for the analysis of drugs. Rapid Commun. Mass Spectrom. 22, 187–196 (2008).
De Spiegeleer, M. et al. Impact of storage conditions on the human stool metabolome and lipidome: preserving the most accurate fingerprint. Anal. Chim. Acta 1108, 79–88 (2020).
Laparre, J. et al. Impact of storage conditions on the urinary metabolomics fingerprint. Anal. Chim. Acta 951, 99–107 (2017).
Cheng, K., Brunius, C., Fristedt, R. & Landberg, R. An LC-QToF MS based method for untargeted metabolomics of human fecal samples. Metabolomics 16, 46 (2020).
Smith, C. A., Want, E. J., O’Maille, G., Abagyan, R. & Siuzdak, G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 78, 779–787 (2006).
Pluskal, T. T., Castillo, S., Villar-Briones, A., Oresic, M. & Orešič, M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinforma. 11, 395 (2010).
Abdi, H. Partial least squares regression and projection on latent structure regression. Wiley Interdiscip. Rev. Comput. Stat. 2, 97–106 (2010).
Bylesjö, M. et al. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J. Chemom. 20, 341–351 (2006).
Quinn, R. A. et al. Microbial, host and xenobiotic diversity in the cystic fibriosis sputum metabolome. ISME J. 10, 1483–1498 (2016).
Smilowitz, J. T. et al. The human milk metabolome reveals diverse oligosaccharide profiles. J. Nutr. 143, 1709–1718 (2013).
Matysik, S., Le Roy, C. I., Liebisch, G. & Claus, S. P. Metabolomics of fecal samples: a practical consideration. Trends Food Sci. Technol. 57, 244–255 (2016).
Naz, S., Vallejo, M., García, A. & Barbas, C. Method validation strategies involved in non-targeted metabolomics. J. Chromatogr. A 1353, 99–105 (2014).
Cameron, S. J. S. et al. Sample preparation free mass spectrometry using Laser-Assisted Rapid Evaporative Ionization Mass Spectrometry: applications to microbiology, metabolic biofluid phenotyping, and food authenticity. J. Am. Soc. Mass Spectrom. 32, 1393–1401 (2021).
Roelants, M., Hauspie, R. & Hoppenbrouwers, K. References for growth and pubertal development from birth to 21 years in Flanders, Belgium. Ann. Hum. Biol. 36, 680–694 (2009).
Cole, T. J. & Lobstein, T. Extended international (IOTF) body mass index cut-offs for thinness, overweight and obesity. Pediatr. Obes. 7, 284–294 (2012).
Zheng, J., Dixon, R. A. & Li, L. Development of isotope labeling LC-MS for human salivary metabolomics and application to profiling metabolome changes associated with mild cognitive impairment. Anal. Chem. 84, 10802–10811 (2012).
Pinto, J. et al. Human plasma stability during handling and storage: impact on NMR metabolomics. Analyst 139, 1168–1177 (2014).
Berkow, S. E. et al. Lipases and lipids in human milk: effect of freeze–thawing and storage. Pediatr. Res. 18, 1257–1262 (1984).
Acknowledgements
This work was supported by VLAIO (grant number IM 150740 – A16/OC/0006), BOF GOA (grant number 2017/000102), BOF (grant number 01J07519), FWO Hercules (grant number AUG/17/09), FWO (grant numbers 1S57920N and 1S49020N) and ERC FWO Runner-up (grant number G0G0119N). The Laboratory of Chemical Analysis research group is part of the Ghent University expertise centre MSsmall.
Author information
Authors and Affiliations
Contributions
V.P., L.V.M., M.D.G., M.D.S., E.D.P. and S.C. wrote the original draft of the manuscript. V.P., L.V.M., E.V.d.W. and A.P. carried out the experiments. M.D.G. and A.P. performed the data analysis. L.V.M., E.D.P. and S.C. collected the samples. Z.T. and S.C. developed the technique. L.V., Z.T. and S.C. supervised the project and provided the funding. L.V., M.D.S. and L.V.M. performed the proofreading and correction of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Arash Zarrine-Afsar and the other, anonymous reviewer(s) for their contribution to the peer review of this work
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Cameron, S. et al. Anal. Chem. 91, 13488 (2019): https://pubs.acs.org/doi/abs/10.1021/acs.analchem.9b02358
Wijnant, K. et al. Anal. Chem. 92, 5116 (2020): https://pubs.acs.org/doi/10.1021/acs.analchem.9b05598
Van Meulebroek, L. et al. Talanta 217, 121043 (2020): https://www.sciencedirect.com/science/article/pii/S0039914020303349
Supplementary information
Supplementary Information
Supplementary Discussion 1.
Rights and permissions
About this article
Cite this article
Plekhova, V., Van Meulebroek, L., De Graeve, M. et al. Rapid ex vivo molecular fingerprinting of biofluids using laser-assisted rapid evaporative ionization mass spectrometry. Nat Protoc 16, 4327–4354 (2021). https://doi.org/10.1038/s41596-021-00580-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-021-00580-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.