Abstract
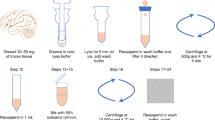
We present a nuclei isolation protocol for genomic and epigenomic interrogation of multiple cell type populations in the human and rodent brain. The nuclei isolation protocol allows cell type-specific profiling of neurons, microglia, oligodendrocytes, and astrocytes, being compatible with fresh and frozen samples obtained from either resected or postmortem brain tissue. This 2-day procedure consists of tissue homogenization with fixation, nuclei extraction, and antibody staining followed by fluorescence-activated nuclei sorting (FANS) and does not require specialized skillsets. Cell type-specific nuclei populations can be used for downstream omic-scale sequencing applications with an emphasis on epigenomic interrogation such as histone modifications, transcription factor binding, chromatin accessibility, and chromosome architecture. The nuclei isolation protocol enables translational examination of archived healthy and diseased brain specimens through utilization of existing medical biorepositories.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The processed ATAC-seq and H3K27ac ChIP-seq tracks for the Schwannoma myeloid cells has been made available as part of a previously published UCSC genome browser session (hg19) containing ATAC-seq, ChIP-seq, and PLAC-seq datasets for brain cell types1: https://genome.ucsc.edu/s/nottalexi/glassLab_BrainCellTypes_hg19. ATAC-seq and ChIP-seq datasets for ex vivo microglia and brain-derived PU.1 nuclei were previously published1,28 and are available on dbGap (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001373.v2.p2).
References
Nott, A. et al. Brain cell type-specific enhancer–promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019).
Breuss, M. W. et al. Somatic mosaicism in the mature brain reveals clonal cellular distributions during cortical development. Preprint at bioRxiv https://doi.org/10.1101/2020.08.10.244814 (2020).
Gallagher, M. D. & Chen-Plotkin, A. S. The post-GWAS era: from association to function. Am. J. Hum. Genet. 102, 717–730 (2018).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Heinz, S., Romanoski, C. E., Benner, C. & Glass, C. K. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 16, 144–154 (2015).
Marzi, S. J. et al. A histone acetylome-wide association study of Alzheimer’s disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci. 21, 1618–1627 (2018).
Nativio, R. et al. An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer’s disease. Nat. Genet. 52, 1024–1035 (2020).
Sun, W. et al. Histone acetylome-wide association study of autism spectrum disorder. Cell 167, 1385–1397.e11 (2016).
Heinz, S. et al. Transcription elongation can affect genome 3D structure. Cell 174, 1522–1536.e22 (2018).
Kempfer, R. & Pombo, A. Methods for mapping 3D chromosome architecture. Nat. Rev. Genet. 21, 207–226 (2020).
Fang, R. et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348 (2016).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Chen, X. et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 13, 1013–1020 (2016).
Wolf, S. A., Boddeke, H. W. G. M. & Kettenmann, H. Microglia in physiology and disease. Annu. Rev. Physiol. 79, 619–643 (2017).
Prinz, M., Jung, S. & Priller, J. Microglia biology: one century of evolving concepts. Cell 179, 292–311 (2019).
Ramamurthy, E. et al. Cell type-specific histone acetylation profiling of Alzheimer’s disease subjects and integration with genetics. Preprint at bioRxiv https://doi.org/10.1101/2020.03.26.010330 (2020).
Hrvatin, S., Deng, F., O’Donnell, C. W., Gifford, D. K. & Melton, D. A. MARIS: method for analyzing RNA following intracellular sorting. PLoS ONE 9, e89459 (2014).
Carlin, A. F. et al. Deconvolution of pro- and antiviral genomic responses in Zika virus-infected and bystander macrophages. Proc. Natl Acad. Sci. USA 115, E9172–e9181 (2018).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Rosenberg, A. B. et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Lu, T. et al. REST and stress resistance in ageing and Alzheimer’s disease. Nature 507, 448–454 (2014).
Siegmund, K. D. et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE 2, e895 (2007).
Spalding, K. L., Bhardwaj, R. D., Buchholz, B. A., Druid, H. & Frisén, J. Retrospective birth dating of cells in humans. Cell 122, 133–143 (2005).
Koshi-Mano, K. et al. Neuron-specific analysis of histone modifications with post-mortem brains. Sci. Rep. 10, 3767 (2020).
van der Poel, M. et al. Transcriptional profiling of human microglia reveals grey–white matter heterogeneity and multiple sclerosis-associated changes. Nat. Commun. 10, 1139 (2019).
Policicchio, S. S. et al. Fluorescence-activated nuclei sorting (FANS) on human post-mortem cortex tissue enabling the isolation of distinct neural cell populations for multiple omic profiling. protocols.io https://doi.org/10.17504/protocols.io.bmh2k38e (2020).
Srinivasan, K. et al. Alzheimer’s patient microglia exhibit enhanced aging and unique transcriptional activation. Cell Rep. 31, 107843 (2020).
Gosselin, D. et al. An environment-dependent transcriptional network specifies human microglia identity. Science 356, eaal3222 (2017).
Nation, D. A. et al. Blood–brain barrier breakdown is an early biomarker of human cognitive dysfunction. Nat. Med. 25, 270–276 (2019).
Sweeney, M. D., Zhao, Z., Montagne, A., Nelson, A. R. & Zlokovic, B. V. Blood–brain barrier: from physiology to disease and back. Physiol. Rev. 99, 21–78 (2019).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017).
Mathys, H. et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380 (2017).
Mathys, H. et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Heinz, S. et al. Effect of natural genetic variation on enhancer selection and function. Nature 503, 487–492 (2013).
Link, V. M. et al. Analysis of genetically diverse macrophages reveals local and domain-wide mechanisms that control transcription factor binding and function. Cell 173, 1796–1809.e17 (2018).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Acknowledgements
We thank M. P. Pasillas for technical assistance and scientific discussions. A.N. was supported by the Alzheimer’s Association (grant no. AARF-18-531498), the Altman Clinical & Translational Research Institute at UCSD (National Center for Advancing Translational Sciences, supported by NIH grant no. KL2TR001444-6), a pilot project grant from UCSD Shiley-Marcos ADRC 1P30AG062429, and the UK Dementia Research Institute, which receives its funding from UK DRI Ltd, funded by the UK Medical Research Council, Alzheimer’s Society, and Alzheimer’s Research UK. J.C.M.S. was supported by the Interdisciplinary Research Fellowship in NeuroAIDS (NIH R25MH081482-12) and the HNRC CSPAR Developmental Core award (NIH 5P30MH062512-18). B.R.F. was supported by the National Institutes of Health (NIH) grant, 1F30AG062159-01. These studies were carried out with grant support to C.K.G. from the NIH R01 NS096170, R01 AG056511, and R01 AG061060-01, and from the Cure Alzheimer’s Fund Gifford Neuroinflammation Consortium CAF 20183159.
Author information
Authors and Affiliations
Contributions
A.N., J.C.M.S., and C.K.G. conceptualized the study; A.N. and J.C.M.S. optimized the methodology with input from B.R.F.; A.N. acquired and analyzed nuclei ChIP-seq and ATAC-seq datasets; A.N., J.C.M.S., B.R.F., and C.K.G. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Dimitrios Davalos, Elvira Mass, and Cindy van Velthoven for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Nott, A. et al. Science 366, 1134−1139 (2019): https://doi.org/10.1126/science.aay0793
Breuss, M. W. et al. Preprint at bioRxiv (2020): https://doi.org/10.1101/2020.08.10.244814
Key data used in this protocol
Nott, A. et al. Science 366, 1134-1139 (2019): https://doi.org/10.1126/science.aay0793
Gosselin, D. et al. Science 356, eaal3222 (2017): https://doi.org/10.1126/science.aal3222
Rights and permissions
About this article
Cite this article
Nott, A., Schlachetzki, J.C.M., Fixsen, B.R. et al. Nuclei isolation of multiple brain cell types for omics interrogation. Nat Protoc 16, 1629–1646 (2021). https://doi.org/10.1038/s41596-020-00472-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00472-3
This article is cited by
-
Quantifying the proportion of different cell types in the human cortex using DNA methylation profiles
BMC Biology (2024)
-
Challenges and opportunities to computationally deconvolve heterogeneous tissue with varying cell sizes using single-cell RNA-sequencing datasets
Genome Biology (2023)
-
Harnessing the potential of machine learning and artificial intelligence for dementia research
Brain Informatics (2023)
-
Neural cell isolation from adult macaques for high-throughput analyses and neurosphere cultures
Nature Protocols (2023)
-
Linking environmental risk factors with epigenetic mechanisms in Parkinson’s disease
npj Parkinson's Disease (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.