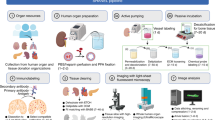
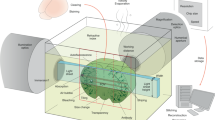
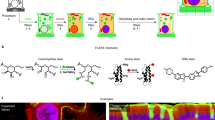
Abstract
Advances in light-sheet and confocal microscopy now allow imaging of cleared large biological tissue samples and enable the 3D appreciation of cell and protein localization in their native organ environment. However, the sample preparations for such imaging are often onerous, and their capability for antigen detection is limited. Here, we describe FLASH (fast light-microscopic analysis of antibody-stained whole organs), a simple, rapid, fully customizable technique for molecular phenotyping of intact tissue volumes. FLASH utilizes non-degradative epitope recovery and membrane solubilization to enable the detection of a multitude of membranous, cytoplasmic and nuclear antigens in whole mouse organs and embryos, human biopsies, organoids and Drosophila. Retrieval and immunolabeling of epithelial markers, an obstacle for previous clearing techniques, can be achieved with FLASH. Upon volumetric imaging, FLASH-processed samples preserve their architecture and integrity and can be paraffin-embedded for subsequent histopathological analysis. The technique can be performed by scientists trained in light microscopy and yields results in <1 week.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
References
Messal, H. A. et al. Tissue curvature and apicobasal mechanical tension imbalance instruct cancer morphogenesis. Nature 566, 126–130 (2019).
Tedeschi, A. et al. Cep55 promotes cytokinesis of neural progenitors but is dispensable for most mammalian cell divisions. Nat. Commun. 11, 1746 (2020).
Huisken, J., Swoger, J., Del Bene, F., Wittbrodt, J. & Stelzer, E. H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009 (2004).
Ueda, H. R. et al. Tissue clearing and its applications in neuroscience. Nat. Rev. Neurosci. 21, 61–79 (2020).
Richardson, D. S. & Lichtman, J. W. Clarifying tissue clearing. Cell 162, 246–257 (2015).
Dekkers, J. F. et al. High-resolution 3D imaging of fixed and cleared organoids. Nat. Protoc. 14, 1756–1771 (2019).
Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606.e19 (2018).
Susaki, E. A. et al. Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging. Nat. Protoc. 10, 1709–1727 (2015).
Susaki, E. A. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739 (2014).
Chung, K. et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337 (2013).
Tainaka, K. et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924 (2014).
Matsumoto, K. et al. Advanced CUBIC tissue clearing for whole-organ cell profiling. Nat. Protoc. 14, 3506–3537 (2019).
Tainaka, K. et al. Chemical landscape for tissue clearing based on hydrophilic reagents. Cell Rep. 24, 2196–2210.e9 (2018).
Murakami, T. C. et al. A three-dimensional single-cell-resolution whole-brain atlas using CUBIC-X expansion microscopy and tissue clearing. Nat. Neurosci. 21, 625–637 (2018).
Cai, R. et al. Panoptic imaging of transparent mice reveals whole-body neuronal projections and skull-meninges connections. Nat. Neurosci. 22, 317–327 (2019).
Cai, R. et al. Panoptic vDISCO imaging reveals neuronal connectivity, remote trauma effects and meningeal vessels in intact transparent mice. Preprint at https://www.biorxiv.org/content/10.1101/374785v1 (2018).
Pan, C. et al. Deep learning reveals cancer metastasis and therapeutic antibody targeting in the entire body. Cell 179, 1661–1676.e19 (2019).
Zhao, S. et al. Cellular and molecular probing of intact human organs. Cell 180, 796–812.e19 (2020).
Chen, B. C. et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998 (2014).
Camacho, D. M., Collins, K. M., Powers, R. K., Costello, J. C. & Collins, J. J. Next-generation machine learning for biological networks. Cell 173, 1581–1592 (2018).
Sullivan, D. P. et al. Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat. Biotechnol. 36, 820–828 (2018).
Wang, H. et al. Deep learning enables cross-modality super-resolution in fluorescence microscopy. Nat. Methods 16, 103–110 (2019).
Todorov, M. I. et al. Machine learning analysis of whole mouse brain vasculature. Nat. Methods 17, 442–449 (2020).
Erturk, A. et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protoc. 7, 1983–1995 (2012).
Erturk, A. et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat. Med. 18, 166–171 (2012).
Yang, B. et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Belle, M. et al. Tridimensional visualization and analysis of early human development. Cell 169, 161–173.e12 (2017).
Murray, E. et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163, 1500–1514 (2015).
Pan, C. et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 13, 859–867 (2016).
Kubota, S. I. et al. Whole-body profiling of cancer metastasis with single-cell resolution. Cell Rep. 20, 236–250 (2017).
Forbes, R. M., Cooper, A. R. & Mitchell, H. H. The composition of the adult human body as determined by chemical analysis. J. Biol. Chem. 203, 359–366 (1953).
McIlwain, H. & Bachelard, H. S. Biochemistry and the central nervous system. (Curchill Livingstone, Edinburgh, UK, 1985).
Shi, S. R., Cote, R. J. & Taylor, C. R. Antigen retrieval techniques: current perspectives. J. Histochem. Cytochem. 49, 931–937 (2001).
Tomer, R., Ye, L., Hsueh, B. & Deisseroth, K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat. Protoc. 9, 1682–1697 (2014).
Tillberg, P. W. et al. Protein-retention expansion microscopy of cells and tissues labeled using standard fluorescent proteins and antibodies. Nat. Biotechnol. 34, 987–992 (2016).
Kim, S. H., Kook, M. C., Shin, Y. K., Park, S. H. & Song, H. G. Evaluation of antigen retrieval buffer systems. J. Mol. Histol. 35, 409–416 (2004).
White, L. J. et al. The impact of detergents on the tissue decellularization process: a ToF-SIMS study. Acta Biomater. 50, 207–219 (2017).
Brito, R. M. & Vaz, W. L. Determination of the critical micelle concentration of surfactants using the fluorescent probe N-phenyl-1-naphthylamine. Anal. Biochem. 152, 250–255 (1986).
Midura, R. J. & Yanagishita, M. Chaotropic solvents increase the critical micellar concentrations of detergents. Anal. Biochem. 228, 318–322 (1995).
Hama, H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat. Neurosci. 14, 1481–1488 (2011).
Ke, M. T., Fujimoto, S. & Imai, T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat. Neurosci. 16, 1154–1161 (2013).
Coutu, D. L., Kokkaliaris, K. D., Kunz, L. & Schroeder, T. Multicolor quantitative confocal imaging cytometry. Nat. Methods 15, 39–46 (2018).
Slaoui, M. & Fiette, L. Histopathology procedures: from tissue sampling to histopathological evaluation. Methods Mol. Biol. 691, 69–82 (2011).
Li, W., Germain, R. N. & Gerner, M. Y. High-dimensional cell-level analysis of tissues with Ce3D multiplex volume imaging. Nat. Protoc. 14, 1708–1733 (2019).
Hamers-Casterman, C. et al. Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 (1993).
Arbabi-Ghahroudi, M. Camelid single-domain antibodies: historical perspective and future outlook. Front. Immunol. 8, 1589 (2017).
Fang, T. et al. Nanobody immunostaining for correlated light and electron microscopy with preservation of ultrastructure. Nat. Methods 15, 1029–1032 (2018).
Susaki, E. A. et al. Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues. Nat. Commun. 11, 1982 (2020).
Hama, H. et al. ScaleS: an optical clearing palette for biological imaging. Nat. Neurosci. 18, 1518–1529 (2015).
Zhao, Y. et al. Nanoscale imaging of clinical specimens using pathology-optimized expansion microscopy. Nat. Biotechnol. 35, 757–764 (2017).
Chen, F., Tillberg, P. & Boyden, E. Expansion microscopy. Science 347, 543–548 (2015).
Shen, K., Sun, J., Cao, X., Zhou, D. & Li, J. Comparison of different buffers for protein extraction from formalin-fixed and paraffin-embedded tissue specimens. PloS One 10, e0142650 (2015).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Horl, D. et al. BigStitcher: reconstructing high-resolution image datasets of cleared and expanded samples. Nat. Methods 16, 870–874 (2019).
Bria, A. & Iannello, G. TeraStitcher—a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics 13, 316 (2012).
Jonkman, J., Brown, C. M., Wright, G. D., Anderson, K. I. & North, A. J. Guidance for quantitative confocal microscopy. Nat. Protoc. Forthcoming (2020).
Smolla, M., Ruchty, M., Nagel, M. & Kleineidam, C. J. Clearing pigmented insect cuticle to investigate small insects’ organs in situ using confocal laser-scanning microscopy (CLSM). Arthropod. Struct. Dev. 43, 175–181 (2014).
Guy, C. T., Cardiff, R. D. & Muller, W. J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 12, 954–961 (1992).
Carlton, J. G. & Martin-Serrano, J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316, 1908–1912 (2007).
Snippert, H. J. et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science 327, 1385–1389 (2010).
Blaas, L. et al. Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat. Cell Biol. 18, 1346–1356 (2016).
Hudry, B. et al. Sex differences in intestinal carbohydrate metabolism promote food intake and sperm maturation. Cell 178, 901–918.e16 (2019).
Acknowledgements
We thank C. Cremona for comments on the manuscript; I. Evans for support with supplies; J. Brock, Research Illustration, for editing of Supplementary Video 7; and the Francis Crick Institute Biological Research, Experimental Histopathology and Advanced Light Microscopy facilities for technical assistance. This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001039), the UK Medical Research Council (FC001039) and the Wellcome Trust (FC001039). This work was also financially supported by the European Molecular Biology Organization (EMBO long-term fellowship ALTF 452-2019 to H.A.M.), the European Research Council (ERC consolidator grant 648804 to J.v.R.), the Doctor Josef Steiner Foundation (to J.v.R.) and the European Research Council (ERC grant 281661 to A.B.).
Author information
Authors and Affiliations
Contributions
H.A.M. conceived and developed the protocol. J.A. contributed to the development of the protocol. H.A.M. and J.A. cowrote the manuscript. M.Z.T. contributed to the comparison with other 3D IF methods and to the development of the adaptation for pigmented tissue and performed vasculature labeling in tumor models. A.T. isolated embryos and embryonic tissues and contributed with the experimental design and analysis of intercellular bridges in 3D. M.Z.T. and A.T. contributed equally. A.C. and K.I.A. provided support with microscopy. L.B. developed the adaptation for insect clearing. I.M.-A., J.v.R. and A.B. supervised the project. All authors read and contributed to the correction of the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.A.M. and A.B. are inventors on a UK patent application (1818567.8) relating to solutions for preparation of samples for 3D imaging.
Additional information
Peer review information Nature Protocols thanks Kazuki Tainaka and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Messal, H. A. et al. Nature 566, 126–130 (2019): https://www.nature.com/articles/s41586-019-0891-2
Tedeschi, A. et al. Nat. Commun. 11, 1746 (2020): https://www.nature.com/articles/s41467-020-15359-w
Extended data
Extended Data Fig. 1 Influence of buffer, pH, temperature and detergent on antigen retrieval.
a, Whole pancreatic lobules were treated for 16 h with the indicated buffers containing 4% (wt/vol) SDS at the indicated temperatures. Shown are representative stainings for Krt19 (pancreatic ducts). Blue lines indicate the minimum temperature above which staining was observed; red lines indicate the maximum temperature above which sample damage was noted. Crosses indicate sample loss. b, 3D view of a pancreatic lobule after cardiac perfusion with dextran-FITC (DexFITC) before and after FLASH reagent 1 treatment (scale bars: 150 µm). c, Comparative whole pancreas immunolabeling for keratin 19 (ducts), C-peptide (islets of Langerhans) and amylase (acinar cells). Samples were incubated at 54 °C with borate alone (left column) and borate with the indicated detergents (all 8% (wt/vol)) without urea. All scale bars: 300 µm.
Extended Data Fig. 2 FLASH reagent 2 preserves the integrity of cytoskeleton and embryos.
a and b, Comparative 3D IF of 100-µm mouse liver sections treated with FLASH reagent 1 or reagent 2. Immunostaining for α-tubulin (tubulin cytoskeleton, a) and cytochrome P450 (Cyt P450) (microsomes, b). Nuclei were stained with DRAQ5. Scale bars: 7 µm (a) and 5 µm (b). c, Images of E13.5 embryos after antigen retrieval with FLASH reagent 1 and reagent 2. d, Comparative IF of 100-µm mouse lung slices treated with FLASH reagent 1 or reagent 2. Immunostaining for α-tubulin and acetylated tubulin (Ac-tubulin; cilia of bronchiolar epithelia). Nuclei were stained with DRAQ5. Scale bars: 50 µm (top panels), 5 µm (center; four panels) and 2 µm (bottom panels).
Extended Data Fig. 3 FLASH compatibility with different RI-matching media.
a, Timeline of sample processing after FLASH staining. b, Representative optical sections showing nuclear staining (Hoechst33342 and DAPI) after RI-matching as indicated. All scale bars: 50 μm. c, Imaging depth for indicated organs and RI-matching media. Note that the tissue depth that can be imaged is not only affected by the optical imaging depth but also by the extent of sample shrinkage/expansion (also see d). A deeper imaging depth of expanded tissue (e.g., in CUBIC-treated pancreas) might capture the same number of cell layers as a lower imaging depth on shrunken tissue. d, Estimate of nuclear density indicating the effect of RI-media on tissue size as shown for the pancreas. e, Signal-to-noise ratio (SNR) for different organs with the indicated RI-media. f, FLASH-compatible mounting media and their approximate RI. BABB, benzyl alcohol/benzyl benzoate; BABB-D4, BABB + diphenyl ether; DBE, dibenzyl ether; DCM (dichloromethane); MetOH, methanol; MetSal, methyl salicylate; tB, tert butanol; TdE, thiodiethanol; THF, tetrahydrofuran.
Extended Data Fig. 4 FLASH with fluorophore-conjugated nanobody staining.
a, Scheme of the alleles in the mouse model with GFP-expressing pancreatic cells used in b and c. b, 3D IF by standard FLASH (primary and secondary antibodies) labeling GFP (pancreatic cells). c, 3D IF of GFP (pancreatic cells) after 1 (left) or 2 (right) d of labeling with a fluorophore-conjugated anti-GFP nanobody. d, Scheme of the alleles in the mouse model with GFP-expressing pancreatic ductal cells used in e and f. e, 3D IF by standard FLASH labeling GFP (pancreatic ductal cells). f, 3D IF of GFP (pancreatic ductal cells) after 1 (left) or 2 (right) d of labeling with a fluorophore-conjugated anti-GFP nanobody. g, Comparison of required time for standard FLASH and FLASH using a fluorophore-conjugated nanobody. Scale bars: 100 µm (b and c) and 50 µm (e and f). AR: antigen retrieval.
Extended Data Fig. 5 Time and clearing comparison in FLASH (reagent 1) and other methods.
a, Comparison of processing time for each of the methods, from tissue collection to mounting for imaging (see also Extended Data Figs. 6–9). b, Images of tissues cleared with different methods. Yellow dotted lines indicate positions of tissue in completely transparent samples. AR: antigen retrieval; Per. or permeab.: permeabilisation; SWITCH: System-Wide control of Interaction Time and kinetics of CHemicals; CUBIC HV: Clear, Unobstructed Brain Imaging Cocktails and Computational analysis HistoVIsion; iDISCO: Immunolabeling-enabled three-Dimensional Imaging of Solvent-Cleared Organs.
Extended Data Fig. 6 3D immunofluorescence of brain cortex with FLASH and other methods.
3D IFs of cortex in 500-µm-thick brain slices with different clearing techniques as indicated, labeling glial fibrillary acidid protein (GFAP; astrocytes) and SMA (vasculature) (a) and TH (axons) (b). Scale bars: 20 µm.
Extended Data Fig. 7 3D immunofluorescence of pancreata with FLASH and other methods.
3D IFs of pancreas with different clearing techniques as indicated, labeling Krt19, TH (nerves) andSMA (vasculature) (a) and Krt19 (ductal cells), C-peptide (C-pep; islets of Langerhans) and Amy (acinar cells) (b). Scale bars: 50 µm.
Extended Data Fig. 8 3D immunofluorescence of mammary glands with FLASH and other methods.
3D IFs of mammary gland with different clearing techniques as indicated, labeling Krt8 (luminal cells), SMA (myoepithelial cells and vasculature) and ColIV (basement membrane) (a); Krt8 and CD31 (endothelial cells) (b); and FoxP1 (nuclei of mammary gland cells) (c). Scale bars: 50 µm (a and b) and 30 µm (c).
Extended Data Fig. 9 Fluorescence intensity and SNR across imaging depth in FLASH and other methods.
a, Stacks of mammary glands imaged after different clearing methods (left and center panels). Look-Up-Tables of Maximum Intensity Projections (MIPs) of the side views show fluorescent intensity across the tissue depth (right panels). Representative images of two independent experiments. Scale bars: 100 µm. b, SNR over the imaging depth of the mammary gland with indicated clearing methods. For CUBIC-HistoVIsion, AbScale and SWITCH, no signal could be observed after up to 200 µm. The signal in FLASH- and iDISCO-treated samples could be discerned through the whole sample thickness.
Supplementary information
Supplementary Information
Supplementary Methods and Supplementary Tables 1–4.
Supplementary Video 1
FLASH of lung. 3D confocal image of bronchiole and arteriole (SMA, magenta), Clara cells (CC10, green) and alveolar type II cells (SFTPC, cyan) in a mouse lung treated with FLASH reagent 1.
Supplementary Video 2
FLASH of pancreas. Confocal image of pancreatic duct (Krt19, white) and islets of Langerhans (PCSK1, red) in a mouse pancreas treated with FLASH reagent 1.
Supplementary Video 3
FLASH of whole mammary gland. Light-sheet image of mammary ducts (Krt8 and Krt5, pink) and basal lamina-defining adipocytes (ColIV, beige) of an entire mammary gland of an adult virgin mouse, treated with FLASH reagent 1.
Supplementary Video 4
FLASH of mammary epithelium. Confocal image of a duct in the mammary gland from Supplementary Video 3, showing luminal cells (Krt8, pink), myoepithelial cells (Krt5, yellow) and basal lamina-defining adipocytes (ColIV, beige).
Supplementary Video 5
FLASH of bile duct. Confocal image of a murine extrahepatic bile duct (Krt19, cyan) and microvasculature (Aqp1, red) treated with FLASH reagent 1.
Supplementary Video 6
FLASH of lacrimal gland. Confocal image of tear ducts (Krt19, green), microvasculature (Aqp1, yellow) and stroma (SMA, magenta) of a mouse lacrimal gland treated with FLASH reagent 1.
Supplementary Video 7
FLASH of wild-type and Cep55−/− embryos. Light-sheet image of whole E13.5 wild-type and Cep55−/− mouse embryos treated with FLASH reagent 2. Outline of entire embryo (autofluorescence, gray) and apoptotic cells (CC3, red). Images of the embryos in this video were included in a previous publication from our laboratory.
Rights and permissions
About this article
Cite this article
Messal, H.A., Almagro, J., Zaw Thin, M. et al. Antigen retrieval and clearing for whole-organ immunofluorescence by FLASH. Nat Protoc 16, 239–262 (2021). https://doi.org/10.1038/s41596-020-00414-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-020-00414-z
This article is cited by
-
Long-term retention of antigens in germinal centers is controlled by the spatial organization of the follicular dendritic cell network
Nature Immunology (2023)
-
Advanced Immunolabeling Method for Optical Volumetric Imaging Reveals Dystrophic Neurites of Dopaminergic Neurons in Alzheimer’s Disease Mouse Brain
Molecular Neurobiology (2023)
-
Antibodies against endogenous retroviruses promote lung cancer immunotherapy
Nature (2023)
-
An analysis modality for vascular structures combining tissue-clearing technology and topological data analysis
Nature Communications (2022)
-
Multispectral confocal 3D imaging of intact healthy and tumor tissue using mLSR-3D
Nature Protocols (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.