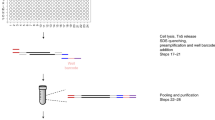
Abstract
Nucleosome organization is important for chromatin compaction and accessibility. Profiling nucleosome positioning genome-wide in single cells provides critical information to understand the cell-to-cell heterogeneity of chromatin states within a cell population. This protocol describes single-cell micrococcal nuclease sequencing (scMNase-seq), a method for detecting genome-wide nucleosome positioning and chromatin accessibility simultaneously from a small number of cells or single cells. To generate scMNase-seq libraries, single cells are isolated by FACS sorting, lysed and digested by MNase. DNA is purified, end-repaired and ligated to Y-shaped adaptors. Following PCR amplification with indexing primers, the subnucleosome-sized (fragments with a length of ≤80 bp) and mononucleosome-sized (fragments with a length between 140 and 180 bp) DNA fragments are recovered and sequenced on Illumina HiSeq platforms. On average, 0.5–1 million unique mapped reads are obtained for each single cell. The mononucleosome-sized DNA fragments precisely define genome-wide nucleosome positions in single cells, while the subnucleosome-sized DNA fragments provide information on chromatin accessibility. Library preparation of scMNase-seq takes only 2 d, requires only standard molecular biology techniques and does not require sophisticated laboratory equipment. Processing of high-throughput sequencing data requires basic bioinformatics skills and uses publicly available bioinformatics software.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data used in this protocol have been deposited in the Gene Expression Omnibus database with accession number GSE96688. Figure 5 is associated with the raw data. There are no restrictions on data availability.
Code availability
Codes used in this protocol have been deposited in GitHub (https://github.com/binbinlai2012/scMNase). There are no restrictions on code availability.
References
Kornberg, R. D. & Lorch, Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98, 285–294 (1999).
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Lai, W. K. M. & Pugh, B. F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 18, 548–562 (2017).
Hodges, C. et al. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325, 626–628 (2009).
Zhang, Z. et al. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332, 977–980 (2011).
Kaplan, N. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366 (2009).
Jiang, C. & Pugh, B. F. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10, 161–172 (2009).
Luger, K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 13, 127–135 (2003).
Lai, B. B. et al. Principles of nucleosome organization revealed by single-cell micrococcal nuclease sequencing. Nature 562, 281–285 (2018).
Schones, D. E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Pott, S. & Lieb, J. D. Single-cell ATAC-seq: strength in numbers. Genome Biol. 16, 172 (2015).
Cooper, J. et al. Genome-wide mapping of DNase I hypersensitive sites in rare cell populations using single-cell DNase sequencing. Nat. Protoc. 12, 2342–2354 (2017).
Jin, W. F. et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature 528, 142–146 (2015).
Baldi, S. Nucleosome positioning and spacing: from genome-wide maps to single arrays. Essays Biochem 63, 5–14 (2019).
Liang, J., Cai, W. & Sun, Z. S. Single-cell sequencing technologies: current and future. J. Genet. Genomics 41, 513–528 (2014).
Hoeijmakers, W. A. M. & Bartfai, R. Characterization of the nucleosome landscape by micrococcal nuclease-sequencing (MNase-seq). Methods Mol. Biol. 1689, 83–101 (2018).
Kelsey, G., Stegle, O. & Reik, W. Single-cell epigenomics: recording the past and predicting the future. Science 358, 69–75 (2017).
Shema, E., Bernstein, B. E. & Buenrostro, J. D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 51, 19–25 (2019).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Pott, S. Simultaneous measurement of chromatin accessibility, DNA methylation, and nucleosome phasing in single cells. eLife 6, 061739 (2017).
Clark, S. J. et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018).
Li, L. et al. Single-cell multi-omics sequencing of human early embryos. Nat. Cell Biol. 20, 847–858 (2018).
Buenrostro, J. D. et al. Transposition of native chromatin for fast and sensitive mulitmodal analysis of chromatin architecture. Biophys. J. 106, 77a (2014).
Buenrostro, J. D. et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Cusanovich, D. A. et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015).
Klein, A. M. & Macosko, E. InDrops and Drop-seq technologies for single-cell sequencing. Lab Chip 17, 2540–2541 (2017).
Perfetto, S. P. et al. Amine-reactive dyes for dead cell discrimination in fixed samples. Curr. Protoc. Cytom. 53, 9.34.1–9.34.14 (2010).
Cui, K. & Zhao, K. Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol. Biol. 833, 413–419 (2012).
Gross, A. et al. Technologies for single-cell isolation. Int. J. Mol. Sci. 16, 16897–16919 (2015).
Kircher, M., Sawyer, S. & Meyer, M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40, e3 (2011).
Langmead, B. & Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Acknowledgements
We thank the National Heart, Lung, and Blood Institute DNA Sequencing Core Facility for sequencing the libraries and the National Heart, Lung, and Blood Institute Flow Cytometry Core facility for sorting the cells. The work was supported by the Division of Intramural Research, National Heart, Lung and Blood Institute (K.Z.), the National Key Research and Development Project (no. 2016YFA0502203) (B.N.), the General Program of National Natural Science Foundation of China (no. 81670534) (B.N.), and the Medical Scientific and Technological Innovation Funds of Southwest Hospital (no. SWH2016LHYS-04) (B.N.).
Author information
Authors and Affiliations
Contributions
K.Z. and B.N. directed the project. W.G. performed the experiments. B.L. analyzed the data. W.G., B.L. and K.Z. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Gonçalo Castelo-Branco, Emily Hodges and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key references using this protocol
Lai, B. et al. Nature 562, 281–285 (2018): https://doi.org/10.1038/s41586-018-0567-3
Integrated supplementary information
Supplementary Fig. 1 FACS sorting of single cells by FSC versus SSC gating.
Formaldehyde-fixed mouse naïve T cells (100,000) were submitted for FACS sorting based on size and granularity (FSC vs SSC, P1 gate), and cell doublets were excluded firstly by SSC-H vs SSC-W (P2 gate) and further by FSC-H vs FSC-W (P3 gate).
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Supplementary Table 1.
Rights and permissions
About this article
Cite this article
Gao, W., Lai, B., Ni, B. et al. Genome-wide profiling of nucleosome position and chromatin accessibility in single cells using scMNase-seq. Nat Protoc 15, 68–85 (2020). https://doi.org/10.1038/s41596-019-0243-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0243-6
This article is cited by
-
Advances in single-cell omics and multiomics for high-resolution molecular profiling
Experimental & Molecular Medicine (2024)
-
New genetic and epigenetic insights into the chemokine system: the latest discoveries aiding progression toward precision medicine
Cellular & Molecular Immunology (2023)
-
Chromatin accessibility profiling by ATAC-seq
Nature Protocols (2022)
-
A guide to systems-level immunomics
Nature Immunology (2022)
-
A plate-based single-cell ATAC-seq workflow for fast and robust profiling of chromatin accessibility
Nature Protocols (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.