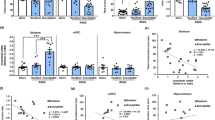
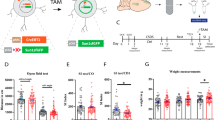
Abstract
Animals susceptible to chronic social defeat stress (CSDS) exhibit depression-related behaviors, with aberrant transcription across several limbic brain regions, most notably in the nucleus accumbens (NAc). Early life stress (ELS) promotes susceptibility to CSDS in adulthood, but associated enduring changes in transcriptional control mechanisms in the NAc have not yet been investigated. In this study, we examined long-lasting changes to histone modifications in the NAc of male and female mice exposed to ELS. Dimethylation of lysine 79 of histone H3 (H3K79me2) and the enzymes (DOT1L and KDM2B) that control this modification are enriched in D2-type medium spiny neurons and are shown to be crucial for the expression of ELS-induced stress susceptibility. We mapped the site-specific regulation of this histone mark genome wide to reveal the transcriptional networks it modulates. Finally, systemic delivery of a small molecule inhibitor of DOT1L reversed ELS-induced behavioral deficits, indicating the clinical relevance of this epigenetic mechanism.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
RNA-seq and ChIP-seq datasets that supported this study (Figs. 4 and 5) have been deposited publicly in the Gene Expression Omnibus under accession code GSE133889. Detailed information on reporting can be found in the linked document titled ‘Life Sciences Reporting Summaryʼ. Source data are provided with this paper.
Code availability
All code used in this work for RNA-seq and ChIP-seq analyses is openly available and can be accessed through the following GitHub repositories, as well as in the file titled ‘Supplementary Software’.
ChIP-seq and RNA-seq raw data processing: https://github.com/shenlab-sinai/NGS-Data-Charmer
ChIP-seq differential analysis: https://github.com/shenlab-sinai/diffreps. Source data are provided with this paper.
Change history
08 April 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41593-021-00848-y
References
Reynolds, S. M. & Berridge, K. C. Emotional environments retune the valence of appetitive versus fearful functions in nucleus accumbens. Nat. Neurosci. 11, 423–425 (2008).
Richard, J. M. & Berridge, K. C. Nucleus accumbens dopamine/glutamate interaction switches modes to generate desire versus dread: D1 alone for appetitive eating but D1 and D2 together for fear. J. Neurosci. 31, 12866–12879 (2011).
Walsh, J. J. et al. Stress and CRF gate neural activation of BDNF in the mesolimbic reward pathway. Nat. Neurosci. 17, 27–29 (2014).
Brancato, A. et al. Sub-chronic variable stress induces sex-specific effects on glutamatergic synapses in the nucleus accumbens. Neuroscience 350, 180–189 (2017).
Lipski, W. J., Dibble, S. M., Rinaman, L. & Grace, A. A. Psychogenic stress activates C-Fos in nucleus accumbens-projecting neurons of the hippocampal ventral subiculum. Int. J. Neuropsychopharmacol. 20, 855–860 (2017).
Gill, K. M. & Grace, A. A. Differential effects of acute and repeated stress on hippocampus and amygdala inputs to the nucleus accumbens shell. Int. J. Neuropsychopharmacol. 16, 2013–2025 (2013).
Cao, J.-L. et al. Mesolimbic dopamine neurons in the brain reward circuit mediate susceptibility to social defeat and antidepressant action. J. Neurosci. 30, 16453–16458 (2010).
Bagot, R. C. et al. Ventral hippocampal afferents to the nucleus accumbens regulate susceptibility to depression. Nat. Commun. 6, 7062 (2015).
Krishnan, V. et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell 131, 391–404 (2007).
Francis, T. et al. Nucleus accumbens medium spiny neuron subtypes mediate depression-related outcomes to social defeat stress. Biol. Psychiatry 77, 212–222 (2014).
Hodes, G. E. et al. Sex differences in nucleus accumbens transcriptome profiles associated with susceptibility versus resilience to subchronic variable stress. J. Neurosci. 35, 16362–16376 (2015).
Dias, C. et al. β-catenin mediates stress resilience through Dicer1/microRNA regulation. Nature 516, 51–55 (2014).
Calipari, E. S. et al. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc. Natl Acad. Sci. USA 113, 2726–2731 (2016).
Bagot, R. C. et al. Circuit-wide transcriptional profiling reveals brain region-specific gene networks regulating depression susceptibility. Neuron 90, 969–983 (2016).
Wook Koo, J. et al. Essential role of mesolimbic brain-derived neurotrophic factor in chronic social stress-induced depressive behaviors. Biol. Psychiatry 80, 469–478 (2016).
Mandelli, L., Petrelli, C. & Serretti, A. The role of specific early trauma in adult depression: a meta-analysis of published literature. Childhood trauma and adult depression. Eur. Psychiatry 30, 665–680 (2015).
Herbison, C. E., Allen, K., Robinson, M., Newnham, J. & Pennell, C. The impact of life stress on adult depression and anxiety is dependent on gender and timing of exposure. Dev. Psychopathol. 29, 1443–1454 (2017).
Delpech, J.-C. et al. Early life stress perturbs the maturation of microglia in the developing hippocampus. Brain. Behav. Immun. 57, 79–93 (2016).
Abbink, M. R., Naninck, E. F. G., Lucassen, P. J. & Korosi, A. Early-life stress diminishes the increase in neurogenesis after exercise in adult female mice. Hippocampus 27, 839–844 (2017).
Hoeijmakers, L. et al. Early-life stress lastingly alters the neuroinflammatory response to amyloid pathology in an Alzheimer’s disease mouse model. Brain. Behav. Immun. 63, 160–175 (2017).
Feifel, A. J., Shair, H. N. & Schmauss, C. Lasting effects of early life stress in mice: interaction of maternal environment and infant genes. Genes Brain Behav. 16, 768–780 (2017).
Wang, X.-D. et al. Forebrain CRF1 modulates early-life stress-programmed cognitive deficits. J. Neurosci. 31, 13625–13634 (2011).
Yajima, H. et al. Early-life stress induces cognitive disorder in middle-aged mice. Neurobiol. Aging 64, 139–146 (2018).
Singh-Taylor, A., Korosi, A., Molet, J., Gunn, B. G. & Baram, T. Z. Synaptic rewiring of stress-sensitive neurons by early-life experience: a mechanism for resilience? Neurobiol. Stress 1, 109–115 (2015).
Yam, K. Y. et al. Exposure to chronic early-life stress lastingly alters the adipose tissue, the leptin system and changes the vulnerability to Western-style diet later in life in mice. Psychoneuroendocrinology 77, 186–195 (2017).
Zhang, Y. et al. Dopamine receptor D2 and associated microRNAs are involved in stress susceptibility and resistance to escitalopram treatment. Int. J. Neuropsychopharmacol. 18, pyv025 (2015).
Peña, C. J. et al. Early life stress confers lifelong stress susceptibility in mice via ventral tegmental area OTX2. Science 356, 1185–1188 (2017).
Lovic, V. et al. Early postnatal experience and DRD2 genotype affect dopamine receptor expression in the rat ventral striatum. Behav. Brain Res. 237, 278–282 (2013).
Romano-López, A. et al. Maternal separation and early stress cause long-lasting effects on dopaminergic and endocannabinergic systems and alters dendritic morphology in the nucleus accumbens and frontal cortex in rats. Dev. Neurobiol. 76, 819–831 (2016).
Peña, C. J. et al. Early life stress alters transcriptomic patterning across reward circuitry in male and female mice. Nat. Commun. 10, 5098 (2019).
Gerfen, C. R. et al. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science 250, 1429–1432 (1990).
Kreitzer, A. C. & Malenka, R. C. Endocannabinoid-mediated rescue of striatal LTD and motor deficits in Parkinson’s disease models. Nature 445, 643–647 (2007).
Chandra, R. et al. Reduced Slc6a15 in nucleus accumbens D2-neurons underlies stress susceptibility. J. Neurosci. 37, 6527–6538 (2017).
Cerdá, M., Sagdeo, A., Johnson, J. & Galea, S. Genetic and environmental influences on psychiatric comorbidity: a systematic review. J. Affect. Disord. 126, 14–38 (2010).
Labonté, B. et al. Sex-specific transcriptional signatures in human depression. Nat. Med. 23, 1102–1111 (2017).
Seney, M. L. et al. Opposite molecular signatures of depression in men and women. Biol. Psychiatry 84, 18–27 (2018).
Kronman, H. et al. Biology and bias in cell type-specific RNAseq of nucleus accumbens medium spiny neurons. Sci. Rep. 9, 8350 (2019).
Reimand, J. et al. g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44, W83–W89 (2016).
Song, L., Pei, L., Yao, S., Wu, Y. & Shang, Y. NLRP3 inflammasome in neurological diseases, from functions to therapies. Front. Cell. Neurosci. 11, 63 (2017).
Alboni, S. et al. Interleukin 18 activates MAPKs and STAT3 but not NF-κB in hippocampal HT-22 cells. Brain. Behav. Immun. 40, 85–94 (2014).
Businaro, R. et al. Interleukin-18 modulation in autism spectrum disorders. J. Neuroinflammation 13, 2 (2016).
Fan, N. et al. Relationship of serum levels of TNF-α, IL-6 and IL-18 and schizophrenia-like symptoms in chronic ketamine abusers. Schizophr. Res. 169, 10–15 (2015).
Vlaming, H. & van Leeuwen, F. The upstreams and downstreams of H3K79 methylation by DOT1L. Chromosoma 125, 593–605 (2016).
Kerry, J. et al. MLL-AF4 spreading identifies binding sites that are distinct from super-enhancers and that govern sensitivity to DOT1L inhibition in leukemia. Cell Rep. 18, 482–495 (2017).
Waters, N. J. Preclinical pharmacokinetics and pharmacodynamics of pinometostat (EPZ-5676), a first-in-class, small molecule S-adenosyl methionine competitive inhibitor of DOT1L. Eur. J. Drug Metab. Pharmacokinet. 42, 891–901 (2017).
Shukla, N. et al. Final report of phase 1 study of the DOT1L inhibitor, pinometostat (EPZ-5676), in children with relapsed or refractory MLL-r acute leukemia. Blood 128, 2780 (2016).
Waters, N. J. et al. Exploring drug delivery for the DOT1L inhibitor pinometostat (EPZ-5676): subcutaneous administration as an alternative to continuous IV infusion, in the pursuit of an epigenetic target. J. Control. Release 220, 758–765 (2015).
Guan, L.-P. & Liu, B.-Y. Antidepressant-like effects and mechanisms of flavonoids and related analogues. Eur. J. Med. Chem. 121, 47–57 (2016).
Arumuggam, N., Bhowmick, N. A. & Rupasinghe, H. P. V. A review: phytochemicals targeting JAK/STAT signaling and IDO expression in cancer. Phyther. Res 29, 805–817 (2015).
Golden, S. A., Covington 3rd, H. E., Berton, O. & Russo, S. J. A standardized protocol for repeated social defeat stress in mice. Nat. Protocols 6, 1183–1191 (2011).
Takahashi, A. et al. Establishment of a repeated social defeat stress model in female mice. Sci. Rep. 7, 12838 (2017).
Sidoli, S., Bhanu, N. V., Karch, K. R., Wang, X. & Garcia, B. A. Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: from histone extraction to data analysis. J. Vis. Exp. 111, 54112 (2016).
Sidoli, S., Simithy, J., Karch, K. R., Kulej, K. & Garcia, B. A. Low resolution data-independent acquisition in an LTQ-Orbitrap allows for simplified and fully untargeted analysis of histone modifications. Anal. Chem. 87, 11448–11454 (2015).
Yuan, Z.-F. et al. EpiProfile 2.0: a computational platform for processing epi-proteomics mass spectrometry data. J. Proteome Res. 17, 2533–2541 (2018).
Cates, H. M. et al. Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol. Psychiatry 84, 167–179 (2018).
Calvanese, V. et al. MLLT3 governs human haematopoietic stem-cell self-renewal and engraftment. Nature 576, 281–286 (2019).
Zhang, X. et al. Prognostic and therapeutic value of disruptor of telomeric silencing-1-like (DOT1L) expression in patients with ovarian cancer. J. Hematol. Oncol. 10, 29 (2017).
Acknowledgements
This work was supported by funding from the National Institutes of Health (P50 MH096890 and R01 MH051399 to E.J.N.) and the Hope for Depression Research Foundation. We also acknowledge R00MH115096 (to C.J.P.), K99DA042100 (to D.M.W.), NARSAD no. 26329 (to O.I.), an Umberto Mortari Award from Merck (to S.S.), grants from the Japan Agency for Medical Research and Development (to A.M.E.D.) and the New York Academy of Sciences (to S.S.).
Author information
Authors and Affiliations
Contributions
H.K., C.J.P., A.T.B., P.M. and E.J.N. designed the studies. C.J.B. and A.T.B. built reversal learning equipment. H.K. and A.T.B. performed all behavioral experiments. With input from B.A.G., S.S. performed mass spectrometry on nuclei isolated by H.K. O.I. and H.K. cloned viral vectors, and R.N. packaged those vectors into viruses. A.G. assisted H.K. with western blotting, E.M.P. assisted H.K. with RNA in situ hybridization and B.B. helped optimize pinometostat administration by running pilot experiments with a separate cohort. H.K. generated all sequencing libraries and performed all qPCRs. C.K.L., Y.V.D.Z. and D.M.W. helped H.K. with stereotactic surgeries and animal sacrifices. A.R., with input from L.S., performed the processing of raw sequencing data as well as the genomic mapping of ChIP-seq data. H.K. performed all downstream data analysis. H.K. wrote the manuscript and prepared all of the figures. All authors discussed the results and edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Peer review information Nature Neuroscience thanks Jeremy Day and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Mass spectrometry data principal components and networks of correlated marks.
(a) Principal component graphs; concentration ellipses determined by means and covariance of groups. Panel A shows PC1/PC2 and Panel B shows PC3/PC4 (b) Correlation by Euclidean distance of histone modifications within group (c) Fold change of epigenetic writers and erasers in male and female post-ELS NAc datasets from Peña et al30.
Extended Data Fig. 2 Viral validation and behavioral specificity.
(a) qPCR of Dot1l in whole NAc tissue following Dot1l knockdown using a cell-type-specific HSV. Significance by two-sided t-test (p=0.02), n=12 individual animals, error bars represent SEM. (b) qPCR of Dot1l in whole NAc tissue following Dot1l overexpression using a cell type-specific HSV. Significance by two-sided t-test (p=0.03), n=13 individual animals, error bars represent SEM. (c) qPCR of Kdm2b in whole NAc tissue following Kdm2b knockdown using a cell type-specific HSV. Significance by two-sided t-test (p=0.01), n=12 individual animals, error bars represent SEM. (d) qPCR of Kdm2b in whole NAc tissue following Kdm2b overexpression using a cell type-specific HSV. Significance by two-sided t-test (p=0.003), n=18 individual animals, error bars represent SEM. (e) Dot1l knockdown in D1 MSNs of male and female mice does not produce social interaction deficits following social defeat. Significance by 2-way ANOVA (n.s.), n=20 individual male animals and 20 individual female animals, error bars represent SEM. (f) Dot1l overexpression in D2 MSNs of the PFC does not produce social interaction deficits following social defeat. Significance by two-sided t-test (n.s.), n=14 individual animals, error bars represent SEM. (g) Snca overexpression in D2 MSNs of the NAc does not produce social interaction deficits following social defeat. Significance by two-sided t-test (n.s.), n=14 individual animals, error bars represent SEM. (h) Social interaction deficits are amplified over the week following social defeat. Significance by two-way ANOVA, n=22 individual animals, error bars represent SEM, lines represent Bonferroni post-test, ** < 0.001, *** < 0.0001.. (i) Std, ELS, and animals with D2 MSN-specific Dot1l overexpression all acquire the initial task with equal accuracy. Significance by two-sided t-test (n.s.), n=29 individual animals, error bars represent SEM. (j) qPCRs of whole NAc tissue from female ELS mice that underwent behavioral testing in Fig. 3. Significance by two-way ANOVA, n=20 individual animals, error bars represent SEM, with lines representing Bonferroni post-test, * < 0.05, ** < 0.001. (k) Correlation of male qPCRs with male behaviors from Figs. 4 and 3, respectively.
Extended Data Fig. 3 Whole tissue detection of H3K79me2 after D2 MSN-specific overexpression of Dot1l.
Performed by ELISA. Significance by two-sided t-test (p=0.03), n=10 individual animals, error bars represent SEM.
Extended Data Fig. 4 ChIP antibody, data quality, and baseline characteristics of H3K79me2 in NAc.
(a) DNA pulled down by IgG and H3K79me2 antibodies. Values represent percent of input (n=2 total samples, each with 2 pooled animals) (b) Distribution of H3K79me2 peaks in Std adult animals (c) Fold enrichment of H3K79me2 peaks in Std adult animals compared to gene length and transcript expression (baseMean value in ELS vs Std DESeq2 comparison). Pearson correlation shows r=-0.15, p<0.00001 (upper panel) and r=-0.01, n.s. (lower panel) (d) Percentage of H3K79me2 peaks in Std adult animals that overlap with enhancer loci predicted in mouse NAc. No enrichment of these enhancer-overlapping peaks.
Extended Data Fig. 5 Pinometostat treatment does not alter locomotor activity or weight.
(a) Schematic of IP Pinometostat administration (b) Locomotor activity after treatment with Pinometostat or saline. Measured in beam breaks. (c) Weight of Pinometostat- and saline-treated animals.
Extended Data Fig. 6 Representative gating for nuclear FACS.
(a) First gate on FSC-A vs SSC-A retrieves nuclei (circled) as opposed to debris (b) Second gate on FSC-A vs Blue1-A (FITC channel) separates transgenically labeled nuclei from wild-type nuclei.
Supplementary information
Supplementary Tables
Worksheet containing Supplementary Tables 1–4 in individual sheets
Source data
Source Data Fig. 2
DOT1L and ACTB full blots, separate stainings, Fig 2b.
Source Data Fig. 6
H3K79me2 and total H3 full blots, separate stainings, Fig 6a.
Rights and permissions
About this article
Cite this article
Kronman, H., Torres-Berrío, A., Sidoli, S. et al. Long-term behavioral and cell-type-specific molecular effects of early life stress are mediated by H3K79me2 dynamics in medium spiny neurons. Nat Neurosci 24, 667–676 (2021). https://doi.org/10.1038/s41593-021-00814-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-021-00814-8
This article is cited by
-
Advancing preclinical chronic stress models to promote therapeutic discovery for human stress disorders
Neuropsychopharmacology (2024)
-
Transcriptomic profiling of the developing brain revealed cell-type and brain-region specificity in a mouse model of prenatal stress
BMC Genomics (2023)
-
Urocortin-3 neurons in the perifornical area are critical mediators of chronic stress on female infant-directed behavior
Molecular Psychiatry (2023)
-
Remembering through the genome: the role of chromatin states in brain functions and diseases
Translational Psychiatry (2023)
-
Epigenetic regulation in major depression and other stress-related disorders: molecular mechanisms, clinical relevance and therapeutic potential
Signal Transduction and Targeted Therapy (2023)