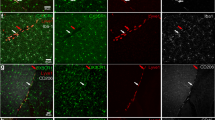
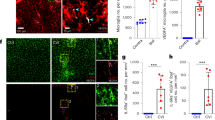
Abstract
Monocyte-derived and tissue-resident macrophages are ontogenetically distinct components of the innate immune system. Assessment of their respective functions in pathology is complicated by changes to the macrophage phenotype during inflammation. Here we find that Cxcr4-CreER enables permanent genetic labeling of hematopoietic stem cells (HSCs) and distinguishes HSC-derived monocytes from microglia and other tissue-resident macrophages. By combining Cxcr4-CreER-mediated lineage tracing with Cxcr4 inhibition or conditional Cxcr4 ablation in photothrombotic stroke, we find that Cxcr4 promotes initial monocyte infiltration and subsequent territorial restriction of monocyte-derived macrophages to infarct tissue. After transient focal ischemia, Cxcr4 deficiency reduces monocyte infiltration and blunts the expression of pattern recognition and defense response genes in monocyte-derived macrophages. This is associated with an altered microglial response and deteriorated outcomes. Thus, Cxcr4 is essential for an innate-immune-system-mediated defense response after cerebral ischemia. We further propose Cxcr4-CreER as a universal tool to study functions of HSC-derived cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Gene expression data are deposited in the GEO database (GSE120701). The data that support the findings of this study and Cxcr4-CreER mice are available from the corresponding authors upon reasonable request.
Code availability
The code for CENA is available at https://github.com/UlasThomas/CoCena2.
References
Alliot, F., Godin, I. & Pessac, B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res. Dev. Brain. Res. 117, 145–152 (1999).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Gomez Perdiguero, E. et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Greter, M., Lelios, I. & Croxford, A. L. Microglia versus myeloid cell nomenclature during brain inflammation. Front. Immunol. 6, 249 (2015).
Parkhurst, C. N. et al. Microglia promote learning-dependent synapse formation through brain-derived neurotrophic factor. Cell 155, 1596–1609 (2013).
Davalos, D. et al. ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758 (2005).
Kettenmann, H., Hanisch, U. K., Noda, M. & Verkhratsky, A. Physiology of microglia. Physiol. Rev. 91, 461–553 (2011).
Prinz, M. & Priller, J. The role of peripheral immune cells in the CNS in steady state and disease. Nat. Neurosci. 20, 136–144 (2017).
Yamasaki, R. et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J. Exp. Med. 211, 1533–1549 (2014).
Ajami, B., Bennett, J. L., Krieger, C., Tetzlaff, W. & Rossi, F. M. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat. Neurosci. 10, 1538–1543 (2007).
Hashimoto, D. et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013).
Kierdorf, K. et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273–280 (2013).
Bowman, R. L. et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 17, 2445–2459 (2016).
Chong, S. Z. et al. CXCR4 identifies transitional bone marrow premonocytes that replenish the mature monocyte pool for peripheral responses. J. Exp. Med. 213, 2293–2314 (2016).
Eash, K. J., Greenbaum, A. M., Gopalan, P. K. & Link, D. C. CXCR2 and CXCR4 antagonistically regulate neutrophil trafficking from murine bone marrow. J. Clin. Invest. 120, 2423–2431 (2010).
Nagasawa, T. CXC chemokine ligand 12 (CXCL12) and its receptor CXCR4. J. Mol. Med. 92, 433–439 (2014).
Nie, Y. et al. The role of CXCR4 in maintaining peripheral B cell compartments and humoral immunity. J. Exp. Med. 200, 1145–1156 (2004).
Sugiyama, T., Kohara, H., Noda, M. & Nagasawa, T. Maintenance of the hematopoietic stem cell pool by CXCL12–CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity 25, 977–988 (2006).
Cruz-Orengo, L. et al. CXCR7 influences leukocyte entry into the CNS parenchyma by controlling abluminal CXCL12 abundance during autoimmunity. J. Exp. Med. 208, 327–339 (2011).
McCandless, E. E., Wang, Q., Woerner, B. M., Harper, J. M. & Klein, R. S. CXCL12 limits inflammation by localizing mononuclear infiltrates to the perivascular space during experimental autoimmune encephalomyelitis. J. Immunol. 177, 8053–8064 (2006).
Banisadr, G. et al. Integrin/chemokine receptor interactions in the pathogenesis of experimental autoimmune encephalomyelitis. J. Neuroimmune Pharmacol. 9, 438–445 (2014).
Ruscher, K. et al. Inhibition of CXCL12 signaling attenuates the postischemic immune response and improves functional recovery after stroke. J. Cereb. Blood Flow Metab. 33, 1225–1234 (2013).
Chia, K., Mazzolini, J., Mione, M. & Sieger, D. Tumor initiating cells induce Cxcr4-mediated infiltration of pro-tumoral macrophages into the brain. eLife 7, e31918 (2018).
Stumm, R. K. et al. A dual role for the SDF-1/CXCR4 chemokine receptor system in adult brain: isoform-selective regulation of SDF-1 expression modulates CXCR4-dependent neuronal plasticity and cerebral leukocyte recruitment after focal ischemia. J. Neurosci. 22, 5865–5878 (2002).
Mass, E. et al. Specification of tissue-resident macrophages during organogenesis. Science 353, aaf4238 (2016).
Stumm, R. K. et al. CXCR4 regulates interneuron migration in the developing neocortex. J. Neurosci. 23, 5123–5130 (2003).
Zou, Y. R., Kottmann, A. H., Kuroda, M., Taniuchi, I. & Littman, D. R. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature 393, 595–599 (1998).
McGrath, K. E., Koniski, A. D., Maltby, K. M., McGann, J. K. & Palis, J. Embryonic expression and function of the chemokine SDF-1 and its receptor, CXCR4. Dev. Biol. 213, 442–456 (1999).
Nie, Y., Han, Y. C. & Zou, Y. R. CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 205, 777–783 (2008).
Roers, A., Hiller, B. & Hornung, V. Recognition of endogenous nucleic acids by the innate immune system. Immunity 44, 739–754 (2016).
Cridland, J. A. et al. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol. Biol. 12, 140 (2012).
Hornung, V., Hartmann, R., Ablasser, A. & Hopfner, K. P. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 14, 521–528 (2014).
Bruttger, J. et al. Genetic cell ablation reveals clusters of local self-renewing microglia in the mammalian central nervous system. Immunity 43, 92–106 (2015).
Kronenberg, G. et al. Distinguishing features of microglia- and monocyte-derived macrophages after stroke. Acta Neuropathol. 135, 551–568 (2018).
Chu, H. X. et al. Evidence That Ly6C(hi) monocytes are protective in acute ischemic stroke by promoting M2 macrophage polarization. Stroke 46, 1929–1937 (2015).
Wattananit, S. et al. Monocyte-derived macrophages contribute to spontaneous long-term functional recovery after stroke in mice. J. Neurosci. 36, 4182–4195 (2016).
Jayaraj, R. L., Azimullah, S., Beiram, R., Jalal, F. Y. & Rosenberg, G. A. Neuroinflammation: friend and foe for ischemic stroke. J. Neuroinflammation 16, 142 (2019).
Ge, R. et al. Choroid plexus-cerebrospinal fluid route for monocyte-derived macrophages after stroke. J. Neuroinflammation 14, 153 (2017).
Jin, W. N. et al. Depletion of microglia exacerbates postischemic inflammation and brain injury. J. Cereb. Blood Flow Metab. 37, 2224–2236 (2017).
Cohen, M. et al. Chronic exposure to TGFβ1 regulates myeloid cell inflammatory response in an IRF7-dependent manner. EMBO J. 33, 2906–2921 (2014).
Kuo, P. C. et al. Interferon-β modulates inflammatory response in cerebral ischemia. J. Am. Heart Assoc. 5, e002610 (2016).
Tran, P. B., Banisadr, G., Ren, D., Chenn, A. & Miller, R. J. Chemokine receptor expression by neural progenitor cells in neurogenic regions of mouse brain. J. Comp. Neurol. 500, 1007–1033 (2007).
Gong, S. et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 (2003).
Nagasawa, T. et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 382, 635–638 (1996).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Watson, B. D., Dietrich, W. D., Busto, R., Wachtel, M. S. & Ginsberg, M. D. Induction of reproducible brain infarction by photochemically initiated thrombosis. Ann. Neurol. 17, 497–504 (1985).
Huang, Z. et al. Effects of cerebral ischemia in mice deficient in neuronal nitric oxide synthase. Science 265, 1883–1885 (1994).
Bederson, J. B. et al. Rat middle cerebral artery occlusion: evaluation of the model and development of a neurologic examination. Stroke 17, 472–476 (1986).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Plescher, M. et al. Plaque-dependent morphological and electrophysiological heterogeneity of microglia in an Alzheimer’s disease mouse model. Glia 66, 1464–1480 (2018).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Amend, S. R., Valkenburg, K. C. & Pienta, K. J. Murine hind limb long bone dissection and bone marrow isolation. J. Vis. Exp. 110, e53936 (2016).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Supek, F., Bosnjak, M., Skunca, N. & Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 (2011).
Bennett, M. L. et al. New tools for studying microglia in the mouse and human CNS. Proc. Natl Acad. Sci. USA 113, E1738–E1746 (2016).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Heng, T. S. et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Acknowledgements
We thank C. Anders, H. Bechmann, S. Bechmann, J. Karius and H. Stadler for excellent technical assistance, and H. Garner and K. Kierdorf for flow cytometry of HSCs. We also thank R. Miller (Northwestern University, Chicago, Illinois, USA) for providing Cxcr4-GFP mice and J. Hansen (German Center for Neurodegenerative Diseases, Bonn, Germany) for providing us with the newest version of MotiQ. This work was supported by German Research Foundation (DFG) grants STU295/3-3 and STU295/8-1 (to R.S.). Work in F.G.’s laboratory (F.G. and E.M.) was supported by grants from the US National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID, 1R01AI130345-01), the National Heart, Lung, and Blood Institute (NIH/NHLBI, 5R01AI124349-03), the National Cancer Institute (NIH/NCI, P30CA08748) and the Ludwig Institute for Cancer Research at MSKCC. E.M. and J.L.S. were funded by the DFG under Germany’s Excellence Strategy EXC2151-390873048 and SCHU 950/8-1 (to J.L.S.). E.M. is supported by the Fritz Thyssen foundation and the Daimler and Benz Foundation.
Author information
Authors and Affiliations
Contributions
E.M., F.G. and R.S. conceived the project. P.A.K., K.H., A.L., E.M., D.S., F.S. and Y.W. performed the experiments. A.H., P.A.K., K.K., E.M., R.S., T.U. and Y.W. analyzed the data. R.S. and Y.W. designed the figures. F.G., E.M., J.L.S. and R.S. supervised experiments and data analyses. C.R. helped establish the stroke models. E.M., F.G. and R.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Paul Kubes and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of Cxcr4-GFP expression.
a, Flow cytometry analysis of GFP signal in postnatal ( > P21) Cxcr4-GFP mice (solid line histograms) and wild-type controls (filled histograms). Cells analyzed were blood leukocytes including B-cells (CD19+), T-cells (CD3+), natural killer cells (NKp46+), neutrophils (Ly6G+), Ly6Clow and Ly6Chi monocytes (CD115+ Ly6Clow / Ly6Chi), CD11b+ F4/80high tissue MΦ (gate 1), and CD11bhigh tissue myeloid cells (gates 2 and 3). Data are representative for n = 4 (blood), n = 7 (spleen, kidney, and liver), and n = 6 (epidermis) mice. b, Confocal micrographs of immunofluorescences for GFP (green) and F4/80 (magenta) in the indicated tissues of postnatal Cxcr4-GFP mice ( ≥ P28). Arrowheads point to Cxcr4-GFP+ F4/80− and arrows to Cxcr4-GFP− F4/80+ cells. Representative for n = 3 mice each. Scale bars: 45 µm (b).
Extended Data Fig. 2 Cxcl12 is dispensable for microglia to colonize the embryonic brain.
a, Confocal analysis of Cxcr4 and Cxcr4-GFP expression in Iba1+ microglia at the indicated embryonic and postnatal stages (n = 1 mouse each). The table shows the number of analyzed Iba1+ cells and the number of co-positives. b, Confocal micrographs demonstrate F4/80/ Cxcr4 and Iba1/ Cxcr4 dual immunofluorescences at E13.5 and E18.5, respectively (images are representative for n = 1 mouse each). c, F4/80 immunofluorescence in coronal head sections of a wild-type mouse and a Cxcl12-/- littermate at E13.5. Graphs depict the number of F4/80+ cells per mm2 determined in 3 matched sectional planes in the dorsal and ventral telencephalon and in one sectional plane in the diencephalon of E13.5 wild-type mice and Cxcl12-/- littermates (images are representative for n = 6 mice each). d, Layering of Iba1+ microglia in cortices of a Cxcr4-deficient mouse and a wild-type littermate at E16.5. The microglia-dense layer to the top represents the subdural meninx (images are representative for n = 4 mice each). The graph shows the frequency of Iba1+ microglia in the indicated cortical layers as percentage of all counted microglia (n = 4 mice each). e, Cxcr4 immunostaining in cortices of a Cxcr4-deficient mouse and a control littermate at E16.5 (n = 4 mice each). Abbreviations: CP, cortical plate; Ctx, cerebral cortex; dTel, dorsal telencephalon; hTh, hypothalamus; IZ, intermediate zone; LV, lateral ventricle; MZ, marginal zone; SVZ, subventricular zone; Th, thalamus; vTel, ventral telencephalon; VZ, ventricular zone. Graphs and statistics: Circles and red lines show individual mice and mean values, respectively. No significant differences as by two-way ANOVA for sectional plane/ genotype interaction for dorsal telencephalon (c, p = 0.06) and ventral telencephalon (c, p = 0.96) and for layer/ genotype interaction for cerebral cortex (d, p = 0.08) and by two-sided t test for diencephalon (c, p = 0.8). Scale bars: 250 µm and 20 µm (b), 500 µm (c), 40 µm (d), and 10 µm (e).
Extended Data Fig. 3 Characterization of Cxcr4CreER/Wt; R26CAG-LSL-tdT mice.
a, Schematic of the Cxcr4-CreER(T2)-IRES-eGFP allele generated by homologous recombination. Upper panel: Cxcr4 exons 1 and 2 (Ex1, Ex2) with the coding sequence in grey. Middle panel: In-frame fusion of the ATG in exon 1 with sequences for CreER(T2), internal ribosomal entry site (IRES), and eGFP. The insertions disrupt the Cxcr4 coding sequence. A neomycin positive selection cassette (neo) flanked by FRT sequences was placed downstream of eGFP. Lower panel: The recombined allele after excision of neo. Primers used for genotyping (arrowheads): Cxcr4-upper (black), Cxcr4-lower (blue), Cxcr4-CreER-lower (purple); primer sequences are detailed in the Methods. Right panel: Southern blot of NheI-digested genomic DNA hybridized with a 5´ Cxcr4 probe identifies the 8.5 kb Flp-excised Cxcr4-CreER fragment and the 4.5 kb Cxcr4 wild-type fragment (blot is representative for n = 7 mice). b, Analysis of tdT signal in tissue MΦ and blood cells in adult mice 4 weeks after TAM. Immunofluorescences show tdT (magenta) and F4/80 (green) in the indicated tissues with DAPI in white. Arrowheads identify tdT+ F4/80−cells and arrows tdT−F4/80+ cells. Images are representative for n = 3 mice. The graph shows flow cytometry analyses of the tdT+ percentage for the indicated cell types (spleen, kidney, and epidermis: n = 4 mice; liver: n = 3 mice). c, Immunostained Cxcr4-IRES-GFP and tdT in TAM-naïve (-TAM) and TAM-treated ( + TAM) P148 mice receiving TAM from P113–P11 (images are representative for n = 3 mice each). d, The graph shows the tdT+ percentage for LSK HSCs (n = 2 mice) and CD11b+ blood cells (n = 5 mice) in adult TAM-naïve (-TAM) condition. e-g, Stroke was induced by tMCAO or PT in adult mice ≥ 28 d after TAM treatment. Micrographs show dual immunofluorescences for tdT/ Iba1 (e, representative for n = 8 mice) and tdT/ Tmem119 (f, representative for n = 8 mice) in the striatal infarct 3 days after tMCAO and a triple immunofluorescence for Cxcr4-IRES-GFP/ tdT/ Iba1 in the cortical infarct 3 days after PT (g, representative for n = 6 mice). DAPI appears white. Graphs depict the tdT+ percentage for Iba1+ cells (e, n = 8 mice) and for GFP+ Iba1+ cells (h, n = 6 mice) as well as the GFP+ percentage for tdT− Iba1+ cells (i, n = 6 mice). Graphs: Circles and red lines represent individual mice and mean values, respectively. Scale bars: 40 µm (b), 500 µm (c), 40 µm (d), 20 µm (e,f), and 15 µm (g).
Extended Data Fig. 4 RNA-seq analysis of the gene expression of monocytes and microglia in photothrombotic infarcts.
a–f,i, RNA-seq was performed with Cxcr4-GFP+ Ly6Chigh monocytes (Ly6G− CD11b+ CD45high) and Cxcr4-GFP− microglia (Ly6G− CD11b+ CD45low) sorted from the injured cortex at day 3 after PT (n = 4 mice). a, Scheme describing the workflow. Differentially expressed (DE) genes were defined by |fold change (FC)| > 4 and false discovery rate (FDR) < 0.3. Statistical test: one-way ANOVA. b, Normalized RNA-seq counts for the indicated genes (center line, median; box limits, 25th to 75th percentiles; whiskers, min to max) (n = 4 mice each). c, Gene Set Enrichment Analysis (GSEA) with all present genes using markers of monocytes, microglia, and neutrophils as gene sets. The plot shows normalized enrichment scores (NES) versus enrichment p-values (FDR). Sources of the marker sets [1] - [3] are detailed in the Methods. Statistical test for nominal p-value of the enrichment score: empirical phenotype-based permutation test. d, Principal component analysis (PCA) with all present genes. e, Hierarchical clustering (HC) showing z-transformed normalized expression values of the top 500 most variable DE genes colored from blue to red. f, GeneOntology (GO) term enrichment analysis of DE genes. Bars depict fold enrichment for terms with p-value < 0.05. Statistical test: EASE Scores (one-tail) calculated with DAVID Bioinformatics. g, h, The indicated transcripts were detected by in situ hybridization (black signal). Images depict the ipsilateral CPu after tMCAO (g) and the border zone of the cortical infarct after PT (h). Infarcts are indicated by asterisks. The contralateral side and the cortex of naïve mice (ctrl) serve as references in (g) and (h), respectively. Sections from mice undergoing tMCAO were counterstained with cresyl violet. Images are representative for n = 2 (ctrl) and n = 3 (stroke). i, Venn diagrams for DE monocyte and microglia genes. The GFP+ monocyte and GFP− microglia gene sets correspond to the data shown in (a). DE genes (|FC| > 1.5 and FDR < 0.05) for tdT+ monocytes and tdT−microglia were obtained by RNA-seq with cells isolated from Cxcr4CreER/Wt; R26CAG-LSL-tdT mice at day 3 after PT (n = 3) and tMCAO (n = 5). Statistical test: Wald test with likelihood ratio test. See Supplementary Table 4 for lists of the DE genes and intersecting sets. Scale bars: 1000 µm (g), 200 µm (h).
Extended Data Fig. 5 Reperfusion facilitates monocyte infiltration in the infarct territory.
a, b, Low magnification images show Cxcr4-GFP at the indicated time points after PT (a) and tMCAO (b) in coronal sections immunostained for GFP and Iba1 (asterisks identify the infarct). Insets represent the framed areas and demonstrate that GFP+ cells in the peri-infarct area are frequently Iba1+ at days 1 and 2 but are Iba1−at day 8. Images are representative for n = 3 mice (day 1 and day 8) and n = 2 mice (day 2) per condition. c–f, High-power confocal images of double-immunofluorescences for GFP/ NeuN (c), GFP/ GFAP (d, e), and GFP/ PECAM (f) at day 3 after stroke induction with GFP in green and NeuN, GFAP, and PECAM in magenta. Cxcr4-GFP+ phagocytes are demonstrated next to NeuN+ neurons (c) and GFAP+ astrocytes (d, e). Few Cxcr4-GFP+ infiltrates are associated with PECAM+ endothelial cells (f, arrowheads). Images are representative for n = 3 mice per condition. g, Dual immunofluorescences for GFP/ Iba1 in the infarct area at day 1 and day 8 after PT and at day 1 after tMCAO. Images are representative for n = 3 mice per condition. Abbreviations: CPu, caudate-putamen; Ctx, cerebral cortex; LV, lateral ventricle. Scale bars: 500 µm (a, b) and 20 µm (c–g).
Extended Data Fig. 6 Effect of Cxcr4cKO on blood leukocytes.
Cxcr4CreER/Wt; R26CAG-LSL-tdT (ctrl) and Cxcr4CreER/LoxP; R26CAG-LSL-tdT mice (cKO) received TAM at adult age and were analyzed ≥ 4 weeks after TAM. a, Relative Cxcr4 expression determined by real-time PCR with cDNA from sorted tdT+ blood monocytes. Single values were normalized to the mean of CreER/Wt. b, Migration index for Cxcl12-induced transwell migration of CD11b+ BM cells. c, d, Numbers of total leukocytes, monocytes (MC), and neutrophilic granulocytes (NG) in gigaparticles (Gpt) per liter blood as determined by leukograms. e, f, Flow cytometry analysis of tdT+ percentages for neutrophilic granulocytes, monocytes, and LSK HSCs. Red lines indicate mean values and squares individual mice. Abbreviations: MC, monocytes; NG, neutrophilic granulocytes. Statistics: a, *p = 0.03 (n = 4 mice each); b, **p = 0.008 (n = 5 mice each); c, p = 0.79 (ctrl, n = 15 mice; cKO, n = 19 mice); d, p = 0.86 (MC), p = 0.07 (NG) (n = 11 mice); e, ***p < 0.0001 (ctrl, n = 10 mice, cKO, n = 9 mice); f, *p = 0.03 (CD150+ CD48−), *p = 0.02 (CD150− CD48−), **p = 0.008 (CD150− CD48+) (n = 5 mice each). a–f, Two-sided Mann-Whitney U test.
Extended Data Fig. 7 Intrainfarct application of AMD3100 induces ectopic positioning of MDM after PT.
Cxcr4CreER/Wt; R26CAG-LSL-tdT mice received saline or AMD3100 into the infarct at day 8 after PT via stereotactically-controlled injection. Images were captured 6 h after substance application. a, Counterstained tdT and GFAP. Regions of interest (ROIs) used to quantify the presence of MDM are illustrated (ROI#1: infarct, ROI#2: peri-infarct, ROI#3: intact region). b, Iba1 staining (green) is shown for cells labeled with arrows in a. DAPI appears white. The depicted tdT+ cells in the saline-treated animal are Iba1− presumptive neurons. The tdT+ Iba1+ cell in the AMD3100-treated animal represents an ectopic MDM in ROI#3. a, b, Images are representative for n = 6 each. Scale bars: 200 µm (a), 20 µm (b).
Extended Data Fig. 8 Diencephalic infarcts and reduced monocyte infiltration in Cxcr4cKO mice undergoing tMCAO.
a, Images demonstrate degenerated areas in the diencephalon of two Cxcr4cKO mice undergoing tMCAO. Segmented lines identify the thalamus and hypothalamus. Arrows point to degenerated/ NeuN−regions. Images are representative for n = 13 mice (day 2) and n = 15 mice (day 3). b, Confocal images of NeuN/ Iba1/ tdT immunofluorescences in a Cxcr4-control (ctrl) and a Cxcr4cKO (cKO) at day 1 after tMCAO. Segmented lines identify the infarct area. High magnification images were photographed in the NeuN+ peri-infarct area in the olfactory tubercle. Images are representative for n = 6 mice each. Abbreviations: CPu, caudate putamen; hTh, hypothalamus; oTb, olfactory tubercle; Th, thalamus. Scale bars: 500 µm (a); 500 µm and 30 µm (b).
Supplementary information
Supplementary Tables
Supplementary Tables 1–6.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 2
Statistical source data
Source Data Fig. 3
Statistical source data
Source Data Fig. 4
Statistical source data
Source Data Fig. 5
Statistical source data
Source Data Fig. 6
Statistical source data
Source Data Fig. 8
Statistical source data
Source Data Extended Data Fig. 2
Statistical source data
Source Data Extended Data Fig. 3
Statistical source data
Source Data Extended Data Fig. 6
Statistical source data
Rights and permissions
About this article
Cite this article
Werner, Y., Mass, E., Ashok Kumar, P. et al. Cxcr4 distinguishes HSC-derived monocytes from microglia and reveals monocyte immune responses to experimental stroke. Nat Neurosci 23, 351–362 (2020). https://doi.org/10.1038/s41593-020-0585-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-0585-y
This article is cited by
-
CD276-dependent efferocytosis by tumor-associated macrophages promotes immune evasion in bladder cancer
Nature Communications (2024)
-
The nuclear factor ID3 endows macrophages with a potent anti-tumour activity
Nature (2024)
-
Mechanism of Efferocytosis in Determining Ischaemic Stroke Resolution—Diving into Microglia/Macrophage Functions and Therapeutic Modality
Molecular Neurobiology (2024)
-
The niche matters: origin, function and fate of CNS-associated macrophages during health and disease
Acta Neuropathologica (2024)
-
Single-Cell Mapping of Brain Myeloid Cell Subsets Reveals Key Transcriptomic Changes Favoring Neuroplasticity after Ischemic Stroke
Neuroscience Bulletin (2024)