Abstract
Maternal infection and inflammation during pregnancy are associated with neurodevelopmental disorders in offspring, but little is understood about the molecular mechanisms underlying this epidemiologic phenomenon. Here, we leveraged single-cell RNA sequencing to profile transcriptional changes in the mouse fetal brain in response to maternal immune activation (MIA) and identified perturbations in cellular pathways associated with mRNA translation, ribosome biogenesis and stress signaling. We found that MIA activates the integrated stress response (ISR) in male, but not female, MIA offspring in an interleukin-17a-dependent manner, which reduced global mRNA translation and altered nascent proteome synthesis. Moreover, blockade of ISR activation prevented the behavioral abnormalities as well as increased cortical neural activity in MIA male offspring. Our data suggest that sex-specific activation of the ISR leads to maternal inflammation-associated neurodevelopmental disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All the sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus with the accession code GSE148237. Publicly available datasets from PANTHER (http://pantherdb.org/) were used for the GO analysis. Source data are provided with this paper.
Code availability
All the custom code are available at https://github.com/bkalishneuro/Maternal-Immune-Activation-Project/blob/main/README.md.
References
Buffington, S. A. et al. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell 165, 1762–1775 (2016).
Hashimoto-Torii, K. et al. Roles of heat shock factor 1 in neuronal response to fetal environmental risks and its relevance to brain disorders. Neuron 82, 560–572 (2014).
Gladwyn-Ng, I. et al. Stress-induced unfolded protein response contributes to Zika virus-associated microcephaly. Nat. Neurosci. 21, 63–71 (2018).
Al-Haddad, B. J. S. et al. Long-term risk of neuropsychiatric disease after exposure to infection in utero. JAMA Psychiatry 76, 594–602 (2019).
Atladóttir, H. O. et al. Maternal infection requiring hospitalization during pregnancy and autism spectrum disorders. J. Autism Dev. Disord. 40, 1423–1430 (2010).
Patterson, P. H. Immune involvement in schizophrenia and autism: etiology, pathology and animal models. Behav. Brain Res. 204, 313–321 (2009).
Lee, B. K. et al. Maternal hospitalization with infection during pregnancy and risk of autism spectrum disorders. Brain. Behav. Immun. 44, 100–105 (2015).
Smith, S. E. P., Li, J., Garbett, K., Mirnics, K. & Patterson, P. H. Maternal immune activation alters fetal brain development through interleukin-6. J. Neurosci. 27, 10695–10702 (2007).
Choi, G. B. et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016).
Shin Yim, Y. et al. Reversing behavioural abnormalities in mice exposed to maternal inflammation. Nature 549, 482–487 (2017).
Zhu, P. J. et al. Activation of the ISR mediates the behavioral and neurophysiological abnormalities in Down syndrome. Science 366, 843–849 (2019).
Larhammar, M. et al. Dual leucine zipper kinase-dependent PERK activation contributes to neuronal degeneration following insult. eLife 6, e20725 (2017).
Green, K. M. et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun. 8, 2005 (2017).
Cheng, W. et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun. 9, 51 (2018).
Loo, L. et al. Single-cell transcriptomic analysis of mouse neocortical development. Nat. Commun. 10, 134 (2019).
Carlezon, W. A. et al. Maternal and early postnatal immune activation produce sex-specific effects on autism-like behaviors and neuroimmune function in mice. Sci. Rep. 9, 16928 (2019).
Gogos, A., Sbisa, A., Witkamp, D. & van den Buuse, M. Sex differences in the effect of maternal immune activation on cognitive and psychosis-like behaviour in Long Evans rats. Eur. J. Neurosci. 52, 2614–2626 (2020).
Rubio Gomez, M. A. & Ibba, M. Aminoacyl-tRNA synthetases. RNA 26, 910–936 (2020).
Evers, R., Smid, A., Rudloff, U., Lottspeich, F. & Grummt, I. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 14, 1248–1256 (1995).
Lessard, F., Stefanovsky, V., Tremblay, M. G. & Moss, T. The cellular abundance of the essential transcription termination factor TTF-I regulates ribosome biogenesis and is determined by MDM2 ubiquitinylation. Nucleic Acids Res. 40, 5357–5367 (2012).
Pitale, P. M., Gorbatyuk, O. & Gorbatyuk, M. Neurodegeneration: keeping ATF4 on a tight leash. Front. Cell. Neurosci. 11, 410 (2017).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Ounallah-Saad, H., Sharma, V., Edry, E. & Rosenblum, K. Genetic or pharmacological reduction of PERK enhances cortical-dependent taste learning. J. Neurosci. 34, 14624–14632 (2014).
Hetz, C. & Papa, F. R. The unfolded protein response and cell fate control. Mol. Cell 69, 169–181 (2018).
Kim, S. et al. Maternal gut bacteria promote neurodevelopmental abnormalities in mouse offspring. Nature 549, 528–532 (2017).
Lammert, C. R. et al. Cutting edge: critical roles for microbiota-mediated regulation of the immune system in a prenatal immune activation model of autism. J. Immunol. 201, 845–850 (2018).
Ryoo, H. D. & Vasudevan, D. Two distinct nodes of translational inhibition in the integrated stress response. BMB Rep. 50, 539–545 (2017).
Rojas, M., Vasconcelos, G. & Dever, T. E. An eIF2α-binding motif in protein phosphatase 1 subunit GADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc. Natl Acad. Sci. USA 112, E3466–E3475 (2015).
McGlincy, N. J. & Ingolia, N. T. Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129 (2017).
Calviello, L. et al. Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods 13, 165–170 (2016).
Merrick, W. C. Eukaryotic protein synthesis: an in vitro analysis. Biochimie 76, 822–830 (1994).
Hoffmann, A. & Roeder, R. G. Cloning and characterization of human TAF20/15. Multiple interactions suggest a central role in TFIID complex formation. J. Biol. Chem. 271, 18194–18202 (1996).
Berson, A. et al. TDP-43 promotes neurodegeneration by impairing chromatin remodeling. Curr. Biol. 27, 3579–3590.e6 (2017).
Marash, L. et al. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell 30, 447–459 (2008).
Henis-Korenblit, S. et al. The caspase-cleaved DAP5 protein supports internal ribosome entry site-mediated translation of death proteins. Proc. Natl Acad. Sci. USA 99, 5400–5405 (2002).
Terenin, I. M., Dmitriev, S. E., Andreev, D. E. & Shatsky, I. N. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 15, 836–841 (2008).
Scheuner, D. et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 7, 1165–1176 (2001).
Sidrauski, C. et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife 2, e00498 (2013).
Sidrauski, C. et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. eLife 4, e07314 (2015).
Sekine, Y. et al. Stress responses. Mutations in a translation initiation factor identify the target of a memory-enhancing compound. Science 348, 1027–1030 (2015).
Chen, Y.-C., Chang, Y.-W. & Huang, Y.-S. Dysregulated translation in neurodevelopmental disorders: an overview of autism-risk genes involved in translation. Dev. Neurobiol. 79, 60–74 (2019).
Alvarez-Castelao, B. & Schuman, E. M. The regulation of synaptic protein turnover. J. Biol. Chem. 290, 28623–28630 (2015).
Costa-Mattioli, M., Sossin, W. S., Klann, E. & Sonenberg, N. Translational control of long-lasting synaptic plasticity and memory. Neuron 61, 10–26 (2009).
Ximerakis, M. et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 22, 1696–1708 (2019).
Lombardo, M. V. et al. Maternal immune activation dysregulation of the fetal brain transcriptome and relevance to the pathophysiology of autism spectrum disorder. Mol. Psychiatry 23, 1001–1013 (2018).
Braun, A. E. et al. “Females are not just ‘protected’ males’: sex-specific vulnerabilities in placenta and brain after prenatal immune disruption. eNeuro https://doi.org/10.1523/ENEURO.0358-19.2019 (2019).
Sutherland, S. & Brunwasser, S. M. Sex differences in vulnerability to prenatal stress: a review of the recent literature. Curr. Psychiatry Rep. 20, 102 (2018).
Guan, B.-J. et al. A unique ISR program determines cellular responses to chronic stress. Mol. Cell 68, 885–900.e6 (2017).
Hrvatin, S. et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129 (2018).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Nygaard, K. R., Maloney, S. E. & Dougherty, J. D. Erroneous inference based on a lack of preference within one group: autism, mice, and the social approach task. Autism Res. 12, 1171–1183 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Ji, Z. RibORF: identifying genome-wide translated open reading frames using ribosome profiling. Curr. Protoc. Mol. Biol. 124, e67 (2018).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Acknowledgements
B.T.K. was supported by the Pediatric Scientist Development Program and the March of Dimes. E.K. was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2018R1A6A3A03010693). M.E.G. and B.T.K. were supported by R01 NS115965 from the National Institute of Neurological Disorders and Stroke. R.J.K. was supported by NIH grants R01CA198103-04, R01DK113171-03 and R01AG062190-02. E.E.D. was supported by the Damon Runyon Foundation. G.B.C. and J.R.H. were supported by the Jeongho Kim Neurodevelopmental Research Fund, the Simons Foundation Autism Research Initiative and National Institute of Mental Health grants (R01MH115037 and R01MH119459, respectively). J.R.H. was also supported by the PEW Scholars Program, the N of One: Autism Research Foundation and the Burroughs Wellcome Fund. Figures 1a, 3b and 4a were created using BioRender (https://biorender.com).
Author information
Authors and Affiliations
Contributions
B.T.K., G.B.C., M.E.G. and J.R.H. conceptualized the study. B.T.K. and E.K. designed and performed the single-cell sequencing experiments. B.T.K., B.F. and E.E.D. performed and analyzed the next-generation sequencing data. E.K. performed the immunoblotting and behavioral analyses. E.K., H.K. and Y.S.Y. bred mice for the experiments. C.K.G., L.T. and B.F. prepared tissues for histology and performed immunohistochemistry and microscopy. B.T.K., E.K., E.C.G., J.R.H. and M.E.G. wrote the manuscript. G.B.C., E.C.G., J.R.H. and M.E.G. provided guidance on the design of experiments and interpretation of results. R.J.K. provided eIF2αS51A mice.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Neuroscience thanks Mauro Costa-Mattioli, John Lukens, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quality Control and E14.5 Sub-clustering.
a, Total number of genes per condition and total number of unique molecular identifiers (UMIs) per condition in the final E14.5 and E18.5 single cell sequencing datasets. Data from n = 2 mice per group. b, UMAP of sub-clustering of mature and immature (SVZ, cortical plate, cortical subplate) neurons at E14.5. Data from n = 2 mice per group. c, UMAP of sub-clustering of radial glia (RG) at E14.5. Data from n = 2 mice per group. d, UMAP of sub-clustering of interneurons (Int), ganglionic eminence (GE), and striatal neurons at E14.5. Data from n = 2 mice per group. e, Dot plot of marker genes associated with all cells at E14.5. Data from n = 2 mice per group. f, Dot plot of marker genes associated with the cells in (b): mature and immature (SVZ, cortical plate, cortical subplate) neurons at E14.5. Data from n = 2 mice per group. g, Dot plot of marker genes associated with the cells in (d): interneurons (Int), ganglionic eminence (GE), and striatal neurons at E14.5. Data from n = 2 mice per group. h, Dot plot of marker genes associated with the cells in (c): radial glia (RG) at E14.5. Data from n = 2 mice per group.
Extended Data Fig. 2 E18.5 sub-clustering.
a, UMAP of sub-clustering of mature and immature (SVZ) neurons at E18.5. Data from n = 2 mice per group. b, UMAP of sub-clustering of radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC) at E18.5. Data from n = 2 mice per group. c, UMAP of sub-clustering of striatal neurons (Str) and interneurons (Int) at E18.5. Data from n = 2 mice per group. d, Dot plot of marker genes associated with cells in (a): mature and immature (SVZ) neurons at E18.5. Data from n = 2 mice per group. e, (left) Dot plot of marker genes associated with cells in (b): radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC) at E18.5; and (right) Dot plot of marker genes associated with cells in (c): striatal neurons (Str) and interneurons (Int) at E18.5. Data from n = 2 mice per group.
Extended Data Fig. 3 Differential gene expression strip plots.
a, Strip plot displaying differential gene expression between MIA male offspring and PBS male offspring at E14.5. Colored dots represent significant genes (FDR < 0.05). The x axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ, cortical plate, cortical subplate) neurons, (2) radial glia (RG), and (3) interneurons (Int), ganglionic eminence (GE), and striatal neurons. Data from n = 2 mice per group. b, Strip plot displaying sex-dependent gene expression in MIA offspring at E14.5. Colored dots represent significant genes (FDR < 0.05). The x axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ, cortical plate, cortical subplate) neurons, (2) radial glia (RG), and (3) interneurons (Int), ganglionic eminence (GE), and striatal neurons. Data from n = 2 mice per group. c, Strip plot displaying differential gene expression between MIA male offspring and PBS male offspring at E18.5. Colored dots represent significant genes (FDR < 0.05). The x axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ) neurons, (2) radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC), and (3) striatal neurons (Str) and interneurons (Int). Data from n = 2 mice per group. d, Strip plot displaying sex-dependent gene expression in MIA offspring at E18.5. Colored dots represent significant genes (FDR < 0.05). The x axis displays select cortical cell types. Cell groups (left to right): (1) mature and immature (SVZ) neurons, (2) radial glia (RG), ganglionic eminence (GE), oligodendrocytes (Olig), and neural stem cells (NSC), and (3) striatal neurons (Str) and interneurons (Int). Data from n = 2 mice per group.
Extended Data Fig. 4 Single cell differential gene expression changes.
a, Bubble plot of highly variable genes between MIA and PBS male offspring at E18.5. All significant genes FDR < 0.05. Blue indicates an increase in MIA males relative to PBS males. Data from n = 2 mice per group. b, Bubble plot of sex-dependent genes in MIA offspring at E18.5. All significant genes FDR < 0.05. Blue indicates an increase in Δ(MIA males – PBS males) relative to females. Data from n = 2 mice per group. c, Bubble plot of ribosome subunit genes between MIA male and PBS male offspring at E14.5, demonstrating a widespread decrease in expression of multiple ribosomal subunits in MIA male offspring. All significant genes FDR < 0.05. Blue indicates an increase in PBS males relative to MIA males. Data from n = 2 mice per group. d, Bubble plot of ribosome subunit genes between MIA female and PBS female offspring at E14.5, demonstrating an increase in expression of multiple ribosomal subunits in MIA female offspring. All significant genes FDR < 0.05. Blue indicates an increase in PBS females relative to MIA females. Data from n = 2 mice per group.
Extended Data Fig. 5 Sex-specific comparisons in control (PBS) conditions.
a, Bubble plot of ribosome subunit genes between PBS male and PBS female offspring at E14.5. All significant genes FDR < 0.05. Data from n = 2 mice per group. b, Bubble plot of ribosome subunit genes between PBS male and PBS female offspring at E18.5. All significant genes FDR < 0.05. Data from n = 2 mice per group.
Extended Data Fig. 6 Upstream regulators of phospho-eIF2α and UPR sensors in E18.5 PBS and MIA fetal cortices.
a, Immunoblot analysis measuring phospho-S6K and quantification in E18 PBS and MIA fetal cortices. The y axis represents relative blot intensity to PBS male control (Two-way ANOVA sex (male or female) × stimulus (PBS or MIA) interaction F1,12 = 0.08413, P = 0.7767; effect of sex F1,12 = 1.188, P = 0.2971; effect of stimulus F1,12 = 0.04729, P = 0.8315 followed by Tukey multiple comparisons test; n = 4 pups from 2 litters). b, Immunoblot analysis measuring upstream regulators of phospho-eIF2α and quantification in E18.5 PBS and MIA fetal cortices. The y axis represents relative blot intensity to PBS control for each sex (Two-tailed unpaired t-test male: pPERK t = 5.485, df = 10, ***P = 0.0003, n = 6 from 3 litters; pGCN2 t = 0.1503, df = 6, n = 4 pups from 2 litters; pPKR t = 0.9501, df = 6, n = 4 pups from 2 litters; Two-tailed unpaired t-test female: pPERK t = 0.5761, df = 6; pGCN2 t = 0.5204, df = 6; pPKR t = 1.545, df = 6; n = 4 pups from 2 litters). c, Immunoblot analysis measuring UPR sensors (IRE1α, ATF6 and PERK) and quantification in E18.5 PBS and MIA male fetal cortices. The y axis represents relative blot intensity to PBS control (Two-tailed unpaired t-test: pIRE t = 2.745, df = 6, *P = 0.0335; ATF6 t = 0.6822, df = 6; pPERK t = 3.516, df = 6, * P = 0.0126; n = 4 pups from 2 litters). Data are mean ± s.e.m.; see Supplementary Table 20 for detailed statistics. Unprocessed blots are provided as a Source Data file.
Extended Data Fig. 7 ISR activation of E18.5 PBS and MIA male fetus in SFB-negative dams.
Immunoblot analysis measuring phospho-eIF2α and quantification in E18.5 PBS and MIA male fetal cortices, either from SFB-positive or SFB-negative B6 dams. SFB-positive dams were obtained from Taconic Biosciences (Tac) whereas SFB-negative dams from Jackson laboratory (Jax) and maintained in SFB-positive and negative conditions, respectively. The y axis represents relative blot intensity to each PBS control (Two-tailed unpaired t-test: Tac t = 4.659, df = 6, **P = 0.0035; Jax t = 0.9013, df = 6; n = 4 pups from 2 litters). Data are mean ± s.e.m.; see Supplementary Table 20 for detailed statistics. Unprocessed blots are provided as a Source Data file.
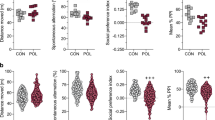
Extended Data Fig. 8 Female MIA offspring neither show ISR activation nor neurodevelopmental abnormalities.
a, Representative images of 8–10 weeks old MIA and PBS female offspring brain tissue immunostained for c-Fos. Scale bar 100 μm. Quantification indicates c-Fos puncta/mm2 (Two-tailed unpaired t-test PBS female versus MIA female: t = 0.1829, df = 5, P = 0.8621; n = 3 for PBS female, n = 4 for MIA female mice). b, Percentage of interaction in the three-chamber sociability test of adult PBS and MIA female offspring littermates (Two-way ANOVA group (PBS female or MIA female) x preference to the target (social target or inanimate) interaction F1,22= 0.986, P = 0.3315; effect of preference to the target F1,22 = 45.24, P = 9.2454 × 10−7 followed by Sidak multiple comparisons test-within group: PBS female ** P = 0.0015, MIA female **** P = 2.06 × 10−5; Two-tailed unpaired t-test social score between PBS female versus MIA female: t = 0.7021, df = 11, P = 0.4972; n = 6 for PBS female, n = 7 for MIA female mice; 2 independent experiments). c, Marble burying index of adult PBS and MIA female offspring littermates (Two-tailed unpaired t-test PBS female versus MIA female: t = 0.9278, df = 15, P = 0.3682; n = 8 for PBS female, n = 9 for MIA female mice; 2 independent experiments). Data are mean ± s.e.m.; see Supplementary Table 20 for detailed statistics.
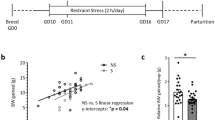
Extended Data Fig. 9 Pharmacological inhibition of ISR protects MIA offspring from neurobehavioral abnormalities.
a, Percentage of interaction in the three-chamber sociability test of vehicle and ISRIB-treated adult PBS and MIA offspring littermates (Two-way ANOVA group (PBS vehicle, MIA vehicle, PBS ISRIB or MIA ISRIB) x preference to the target (social target or inanimate) interaction F3,62 = 7.401, P = 0.003; effect of preference to the target F1,62 = 57.62, P = 2 × 10−10 followed by Bonferroni multiple comparisons test-within group: PBS vehicle **** P = 4.9 × 10−10, PBS ISRIB * P = 0.0119, MIA vehicle P > 0.9999, MIA ISRIB **** P = 2.76 × 10−5; two-way ANOVA stimulus (PBS or MIA) x treatment (vehicle or ISRIB) interaction F1,31 = 7.111, P = 0.0121; effect of stimulus F1,31 = 4.109, P = 0.0513; effect of treatment F1,31 = 0.003618, P = 0.9524 followed by Dunnett multiple comparisons test: PBS vehicle versus PBS ISRIB P = 0.19, MIA vehicle versus PBS vehicle **P = 0.0071, PBS vehicle versus MIA ISRIB P = 0.3031; n = 10 for PBS vehicle, n = 7 for MIA vehicle, n = 7 for PBS ISRIB, n = 11 for MIA ISRIB; 2 independent experiments). b, Marble burying index of vehicle and ISRIB-treated adult PBS and MIA offspring littermates (Two-way ANOVA stimulus (PBS or MIA) x treatment (vehicle or ISRIB) interaction F1,13 = 4.549, P = 0.0526; effect of stimulus F1,13 = 8.829, P = 0.0108; effect of treatment F1,13 = 4.341, P = 0.0575 followed by Tukey multiple comparisons test: PBS vehicle versus MIA vehicle *P = 0.0147, PBS ISRIB versus MIA vehicle *P = 0.0255, MIA vehicle vs. MIA ISRIB *P = 0.0275; n = 4 for PBS vehicle, n = 4 for MIA vehicle, n = 3 for PBS ISRIB, n = 6 for MIA ISRIB mice; 2 independent experiments). Data are mean ± s.e.m.; see Supplementary Table 20 for detailed statistics.
Supplementary information
Supplementary Table 1
DGE between MIA male and PBS male offspring in E14 scRNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(counts per million) (log(CPM)), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 2
Supplementary Table 2 DGE between MIA female and PBS female offspring in E14 scRNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 3
DGE between PBS male and PBS female offspring in E14 scRNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 4
DGE between MIA male and MIA female offspring in E14 scRNA-seq using interaction term analysis. Data analyzed using edgeR with quasi-likelihood F test. Data include log2 (FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 5
DGE between MIA male and PBS male offspring in E18 scRNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 6
DGE between MIA female and PBS female offspring in E18 scRNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 7
DGE between PBS male and PBS female offspring in E18 scRNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 8
DGE between MIA male and MIA female offspring in E18 scRNA-seq using interaction term analysis. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 9
Quality control metrics for ribosome-profiling data.
Supplementary Table 10
DGE between MIA male and PBS male offspring in E18 bulk RNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 11
DGE between MIA female and PBS female offspring in E18 bulk RNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2 (FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 12
DGE between PBS male and PBS female offspring in E18 bulk RNA-seq. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 13
DGE between MIA male and MIA female offspring in E18 bulk RNA-seq using interaction term analysis. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 14
DGE between MIA male and MIA female offspring in E18 bulk RNA-seq using a pairwise comparison. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 15
Differential translation analysis between MIA male and PBS male offspring in E18 ribosome profiling. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 16
Differential translation analysis between MIA female and PBS female offspring in E18 ribosome profiling. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM, F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 17
Differential translation analysis between PBS male and PBS female offspring in E18 ribosome profiling. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 18
Differential translation analysis between MIA male and MIA female offspring in E18 ribosome profiling using interaction term analysis. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 19
Differential translation analysis between MIA male and MIA female offspring in E18 ribosome profiling using a pairwise comparison. Data analyzed using edgeR with quasi-likelihood F test. Data include log2(FC), log(CPM), F statistics, P values (unadjusted) and FDR-adjusted values for multiple comparisons.
Supplementary Table 20
Supporting statistical data for Figs. 3, 5 and 6 and Extended Data Figs. 6–9.
Supplementary Table 21
Supporting data for Fig. 4c.
Source data
Source Data Fig. 3
Uncropped western blots.
Source Data Fig. 5
Uncropped western blots.
Source Data Extended Data Fig. 6
Uncropped western blots.
Source Data Extended Data Fig. 7
Uncropped western blots.
Rights and permissions
About this article
Cite this article
Kalish, B.T., Kim, E., Finander, B. et al. Maternal immune activation in mice disrupts proteostasis in the fetal brain. Nat Neurosci 24, 204–213 (2021). https://doi.org/10.1038/s41593-020-00762-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-020-00762-9
This article is cited by
-
Chorioamnionitis accelerates granule cell and oligodendrocyte maturation in the cerebellum of preterm nonhuman primates
Journal of Neuroinflammation (2024)
-
Prenatal and postnatal neuroimmune interactions in neurodevelopmental disorders
Nature Immunology (2024)
-
Human brain organoid model of maternal immune activation identifies radial glia cells as selectively vulnerable
Molecular Psychiatry (2023)
-
Infection of the murine placenta by Listeria monocytogenes induces sex-specific responses in the fetal brain
Pediatric Research (2023)
-
An old model with new insights: endogenous retroviruses drive the evolvement toward ASD susceptibility and hijack transcription machinery during development
Molecular Psychiatry (2023)