Abstract
The twenty-first century has already recorded more than ten major epidemic or pandemic virus emergence events, including the ongoing and devastating coronavirus disease 2019 (COVID-19) pandemic. As viral disease emergence is expected to accelerate, these data dictate a need for proactive approaches to develop broadly active family-specific and cross-family therapeutics for use in future disease outbreaks. Emphasis should focus not only on the development of broad-spectrum small-molecule and antibody direct-acting antivirals, but also on host-factor therapeutics, including repurposing previously approved or in-pipeline drugs. Another new class of therapeutics with great antiviral therapeutic potential is RNA-based therapeutics. Rather than only focusing on known risks, dedicated efforts must be made toward pre-emptive research focused on outbreak-prone virus families, ultimately offering a strategy to shorten the gap between outbreak and response. Emphasis should also focus on orally available drugs for outpatient use, if possible, and on identifying combination therapies that combat viral and immune-mediated pathologies, extend the effectiveness of therapeutic windows and reduce drug resistance. While such an undertaking will require new vision, dedicated funding and private, federal and academic partnerships, this approach offers hope that global populations need never experience future pandemics such as COVID-19.
Similar content being viewed by others
Main
In December 2019, a novel virus was identified in a cluster of human cases in Wuhan, China. The virus, identified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2; a coronavirus related to the virus that caused the 2003 SARS epidemic), quickly spread throughout China and then globally1. COVID-19, the disease caused by SARS-CoV-2, has raged across the world, causing more than 100 million reported cases and greater than 2 million deaths. As of early 2021, the COVID-19 pandemic is still ongoing, causing enormous economic losses, closing of uncountable numbers of businesses and loss of employment for millions. As with the deadly 1918 influenza outbreak almost exactly 100 years prior, the world was unprepared for SARS-CoV-2—despite the 2003 SARS-CoV pandemic, an ongoing Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak since 2012 and warnings of high-risk coronavirus strains circulating in bats2. In the first months of the COVID-19 pandemic, initial responses included public health measures such as business closings, stay-at-home orders, mask wearing and social distancing, which were effective against earlier outbreaks. However, SARS-CoV-2 was capable of community and asymptomatic spread, circumventing classic control strategies in most nations globally. Revolutionary advances in fundamental virology, immunology, biochemistry and cell biology research led to novel vaccines and antiviral products in record time, yet distribution of these products to 7 billion individuals across the globe remains a daunting challenge. In parallel, the rapid spread of misinformation on social media continues to sow confusion and erode public confidence in medical interventions and heighten anxiety and confusion.
In the year since SARS-CoV-2 emerged, the scientific community has made incredible progress in developing therapeutic regimens. One of the most well-known SARS-CoV-2 antivirals is remdesivir, previously shown to be a potent inhibitor of a panel of contemporary epidemic and zoonotic coronaviruses both in vitro and in vivo3. It was quickly shown that remdesivir also possessed robust antiviral activity against SARS-CoV-2 (ref. 4), and successful clinical trials rapidly led to its approval for emergency use by the Food and Drug Administration (FDA) in May 2020 in the USA and elsewhere. In August, convalescent plasma from individuals who have recovered from COVID-19 was approved for use in the USA under emergency authorization. The European Medicines Agency endorsed use of the corticosteroid dexamethasone for late-stage COVID-19 disease in September5. Soon thereafter, in November, the FDA approved the arthritis drug baricitinib (in combination with remdesivir) for emergency use. Two novel monoclonal antibody therapeutics were approved for COVID-19 emergency use by the FDA in November of 2020: bamlanivimab6 and a combination of casirivimab and imdevimab7. Thus far, these therapeutics have been associated with reduced viral load in patients, reduced time to recovery and/or reduced progression to severe disease5,6,7,8,9. Although a great success, these novel therapeutics were not developed, tested and approved until nearly a full year after the outbreak emerged. More recently, the first broadly cross-neutralizing human monoclonal antibodies have been described that target many different sarbecoviruses10. Finally, in December 2020, the adenovirus-based AstraZeneca–Oxford vaccine and two mRNA-based vaccines (Pfizer–BioNTech and Moderna) were approved for COVID-19 emergency use in England, the USA and elsewhere11,12,13. A wide portfolio of repurposed drugs, therapeutic antibodies and antiviral drugs are currently in clinical trials, providing novel opportunities for identifying SARS-CoV-2 and broadly active pan-coronavirus intervention strategies.
The SARS-CoV-2 pandemic has highlighted a critical need for investment in preparedness for global outbreaks by infectious agents of the future, including systemic investment in public health preparedness, diagnostics and intervention technologies. As highlighted by the cases of remdesivir3, molnupiravir (a nucleoside analog developed for influenza and repurposed for SARS-CoV-2)4 and the human monoclonal antibody ADG2 (an antibody therapeutic active against multiple coronavirus family members)14, it is vital to develop and identify family-wide or group-specific therapeutics and vaccines that can treat highly heterogeneous unknown zoonotic viruses that may emerge in the future. By having broad-based drugs on the shelf that can be tested immediately in clinical trials and rapidly distributed under emergency use authorization early in an expanding pandemic, we can delay spread, attenuate virus evolution and adaptation to the new human host, and narrow the window between emergence and response. This review will look back on twenty-first-century outbreaks with an emphasis on COVID-19 to discuss high-risk, zoonotic RNA virus pathogens with epidemic and pandemic potential. We will consider different classes of therapeutics, discuss the possibility of repurposing existing therapeutics and suggest a structure to stimulate pre-emptive therapeutic development required to change global preparedness patterns from a reactive to proactive strategy.
Recent outbreaks and potential threats
In the last two decades of the twenty-first century, there have been more than ten major viral disease epidemics or pandemics in human populations, caused by coronavirus, alphavirus, myxovirus, filovirus, norovirus and flavivirus family members (Fig. 1). In addition, small sporadic outbreaks have originated from henipaviruses, bunyaviruses, arenaviruses and other zoonotic RNA viruses. In parallel, zoonotic viruses frequently spillover into economically important livestock and other animals, which can serve as reservoir hosts for spillover into humans, demonstrating the critical importance of a one-health strategy designed to unite human and veterinary medicine and public health practices to control future threats. Unfortunately, it is difficult to predict future outbreaks beyond recognizing the existence of pre-epidemic forms that circulate in reservoirs and appear to spillover sporadically into mammalian and human hosts2,15,16.
Coronaviruses
One of the first outbreaks of the twenty-first century was the SARS outbreak, caused by SARS-CoV. SARS-CoV emerged in late 2002 and quickly spread around the world before containment by the summer of 2003. There were several thousand known cases during this time, with a fatality rate of roughly 10%17. In 2012, another novel zoonotic coronavirus, MERS-CoV, emerged to cause MERS. Sporadic outbreaks and cases of MERS have continued since 2012. Although the total number of MERS-CoV cases is lower, the mortality rate is substantially higher, approaching 35%18. Finally, SARS-CoV-2 emerged in 2019, causing the current COVID-19 pandemic. All of these coronaviruses were traced back to bat origin, some using intermediate hosts such as camels and civet cats19. Moreover, there are a number of pre-epidemic group I and II coronaviruses circulating in bats that can replicate in primary human cells and are poised for emergence15,20.
Flaviviruses
Mosquito-borne flaviviruses, including dengue virus (DENV), Zika virus (ZIKV), West Nile virus and yellow fever virus (YFV), are novel or re-emerging pathogens defined as category A/B pathogens by the National Institutes of Health (NIH; https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens). DENV is endemic to areas of southeast Asia and the Americas, punctuated every few years by the appearance of novel strains that have caused major epidemics since the early 2000s21. The Dengvaxia vaccine is licensed in multiple countries for individuals who have previously had a DENV infection, although it is contraindicated in naive individuals22. In 2015, ZIKV arrived from Polynesia and emerged in Brazil, quickly sweeping throughout South and Central America and the Caribbean. Although mortality rates were low, devastating cases of microcephaly and other central nervous system abnormalities were identified in newborn infants from mothers infected early in pregnancy21. In 2016, YFV re-emerged in Angola, spreading to neighboring countries in Africa and then to China through travel, before being controlled by mass vaccination21. These flaviviruses are mosquito-borne, and the range of disease is expected to increase as a consequence of global warming, travel and spread of vectors across the globe23. Multiple tick-borne flaviviruses also exist, representing potential public health threats.
Influenza viruses
Influenza viruses have caused several well-characterized outbreaks in the twentieth century, including the devastating 1918 H1N1 flu pandemic, with about 50 million deaths globally. Other epidemics were recorded in 1957 (H2N2 Asian flu) and 1968 (H3N2 Hong Kong flu)24, as well as sporadic human cases of highly pathogenic avian flu strains. In the twenty-first century, the 2009 H1N1 swine flu pandemic strain emerged, causing about 250,000 deaths globally25. This strain, related to 1918 H1N1, originated in pigs and was a triple reassortment of avian, swine and human influenza strains26. The twenty-first century has seen continued sporadic outbreaks of avian H5N1, H7N9 and other influenza strains, which have very high mortality rates in human cases but fortunately have limited human-to-human transmissibility27. Major research campaigns are underway to develop universal vaccines and drugs for influenza viruses28.
Filoviruses
The filoviruses Ebola virus (EBOV) and Marburg virus (MARV) are defined as category A pathogens by the NIH (https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens) and the Centers for Disease Control and Prevention (CDC; https://emergency.cdc.gov/agent/agentlist-category.asp). There were two major EBOV outbreaks in the last decade. The 2013–2016 epidemic was primarily in West African countries, infecting ~30,000 individuals with a mortality rate of 40%29. There was a second large outbreak in the Democratic Republic of the Congo from 2018 to 2020, with around 3,500 infections and a mortality rate of 65%30. A highly efficacious vaccine candidate was approved by the World Health Organization in 2019 and more than 200,000 people were vaccinated31, helping to control the outbreak, although vaccination efforts were hampered by armed conflict in the region. Bats are the primary reservoir for EBOV, and virus can transmit either directly to humans or through intermediate zoonotic hosts. Once in the human population, EBOV can spread through contact with blood and bodily fluids, or through sexual transmission25.
Alphaviruses
The alphavirus chikungunya virus (CHIKV) is naturally found in Africa and Asia. In 2006, there was a large outbreak in India, with around 1 million suspected cases32. The virus then spread to the Americas, causing large epidemics between 2013 and 2016 with several million cases. Like the flaviviruses discussed above, CHIKV is a mosquito-borne virus, and thus restricted in range, although this is predicted to change owing to vector spread and global climate change. Although rarely fatal, CHIKV can cause long-term debilitating chronic disease after primary infection33. Additionally, there is a pool of other alphavirus family members (Venezuelan equine encephalitis virus (VEEV), Eastern equine encephalitis virus, Mayaro virus and so on) that represent a real and growing threat to the health of communities and domesticated animals and are category B pathogens on NIH and CDC lists34,35.
Other viruses of concern
The order Bunyavirales contains multiple viruses that have caused outbreaks or have outbreak potential, including Rift Valley fever virus (RVFV), Lassa fever virus (LASV), Crimean–Congo hemorrhagic fever virus (CCHFV) and hantaviruses. These viruses are category A priority pathogens on the NIH (https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens) and CDC (https://emergency.cdc.gov/agent/agentlist-category.asp) lists owing to their high mortality rates (10–40%) and possibility of transmission through insect and rodent vectors36,37,38,39. There have been multiple small outbreaks of henipaviruses, including a 2018 Nipah virus (NiV) outbreak in India40, with high mortality rates (60–90%). Bats are the reservoir of henipaviruses, and intermediate hosts include pigs and horses40,41; an equine vaccine has been developed for Hendra virus (HeV) to help limit transmission to humans42. Norovirus is the leading cause of gastrointestinal infections, triggering multiple epidemics in the last two decades leading to severe disease in children, the elderly and immunocompromised individuals and a projected 70,000–200,000 deaths per year globally. Norovirus is extremely diverse, which may hamper development of vaccines and antivirals43,44. Other viruses considered high priority by the NIH include hepatitis A and rabies virus (https://www.niaid.nih.gov/research/emerging-infectious-diseases-pathogens).
Development of critical tools and methods
The major roadblocks impeding the development of broadly active antiviral therapeutics include targeting therapies against an unknown pathogen, the lack of relevant models for in vitro and in vivo screening, unknown market potential, risk aversion associated with drug repurposing and prioritization of scarce resources for pandemic preparedness.
Viral isolates and molecular clones
RNA virus families are highly heterogeneous, but metagenomic and viral discovery efforts have identified full-length genome sequences representing many genetically diverse viral branches across the family tree45, providing opportunities to select disparate, genetically diverse strains for drug and vaccine testing. As pandemic strains of the future are unknown, drugs that robustly block diverse virus family members will increase confidence for activity against an unknown. Sometimes, with difficulty, a virus can be isolated from a clinical, animal or environmental sample in cell culture or animals46. More recently, synthetically derived molecular clones of the virus genome can be generated through reverse genetics using known viral sequences, providing pure stocks of viruses representing different phylogenetic branches47,48,49. Moreover, synthetic biology offers opportunities for the creation of reporter viruses, which are extremely helpful in the development of high-throughput workflows50,51. This approach allows systematic screening of diverse zoonotic strains designed to identify pre-epidemic strains that are capable of replicating in human primary cells. Although a variety of recombinant protein biochemical assays can be used for high-throughput drug screens, the development of remdesivir, molnupiravir and the human monoclonal antibody ADG2 (mentioned above) utilized a combination of contemporary human coronaviruses, emerging coronaviruses and pre-epidemic high- and low-risk zoonotic coronaviruses, derived by reverse genetics, chosen to enhance confidence that the therapeutics will work against an unknown strain.
Cell culture models of viral infection
Along with a portfolio of viruses, it is necessary to have robust model systems that reflect the relevant human tissues. Culturing viruses from samples can be a difficult task; use of innate immunodeficient cells or receptor-expressing cells (for viruses with known receptors) enhances the likelihood of successful culture49. High-throughput screening methods are necessary to reduce the time, manpower and expense of novel therapeutic development. As a result of availability and expense, it is common to use transformed human cell lines that can be infected by the target virus for high-throughput screens. Although this approach has led to identification of successful drugs, these transformed cell lines often lack relevance, affecting drug bioavailability, transport and metabolism. Portfolios of primary cell lines are also available, informing drug bioavailability in multiple susceptible cell types that may contribute to pathology and disease in humans49,52. Major advances in human organ-on-a-chip and stem cell research have also occurred over the past two decades, affording relevant primary human tissues from different organs that are infected by RNA viruses53,54,55. For greatest efficacy, initial screening should be carried out in primary cell lines; greater expense and effort will be offset by increased relevance and a greater likelihood that hits will be successful, especially for host-targeted therapeutics. After primary hit identification, candidates should be tested in organ-on-a-chip systems for further validation before evaluation in animal models of disease. Screening efforts will be greatly benefited by collaborations to pool expertise in virology, culture models and screening methods, and share the burden of large-scale drug testing.
Animal models of viral disease
Small-animal models are the next step for screening of drug efficacy. Differences in genetics and expression patterns in specific species can complicate data interpretation, including the problem of comparative biology in outbred humans versus inbred models. However, small- and large-animal models remain important in therapeutic development to recapitulate an organismal response to infection. The most common animal model for research is the laboratory mouse, although ferrets, guinea pigs and hamsters are also commonly used for studying viral pathogenesis and transmission or to achieve the FDA two-animal requirement. It is critical that mouse models recapitulate high virus titers and many of the severe disease phenotypes that occur in humans. However, not all viruses of interest will naturally replicate efficiently or even infect laboratory animals, and new models may be needed. For mice, one approach is to use genetically diverse inbred populations such as the Collaborative Cross that capture the genetic diversity of Mus musculus and may display severe disease that phenocopies human disease56. If the viral receptor is known, mouse models can be created that express the human receptor. This can be achieved in multiple ways, including through viral transduction57 or creation of stable receptor-expressing mice through genetic techniques including CRISPR–Cas9 (ref. 58). Knock-in models have the advantage of using mouse regulatory loci to ensure host gene expression patterns, while transgenic mice often express receptor in irrelevant tissues, hampering data interpretation.
An alternate approach is to select for more virulent viruses in the alternative host, rather than engineering susceptibility into the animal. Although some viruses replicate efficiently in mice, the animals may not display disease symptoms like those seen in human infections. Using experimental evolution, virus can be serially passaged in mice or other laboratory animals to generate a more pathogenic variant58. Although powerful, one possible drawback to this approach is that by introducing alterations into the virus, antigenicity or viral response to therapeutics may be affected. Nevertheless, it is preferable to use models that recapitulate human disease phenotypes and tissue tropism to minimize complications in data interpretation across species.
Therapeutic types and mechanisms of action
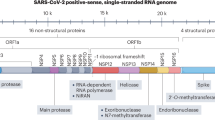
Although each virus has distinct biology underlying its infection, they all share several fundamental steps that represent targets for therapeutic development (Fig. 2a). The first step in viral infection is viral entry. Proteins on the viral exterior (capsid or envelope) bind and interact with specific receptors or attachment factors, which can be proteins, glycans and/or lipids, such as the SARS-CoV-2 host receptor ACE2 (ref. 59). These interactions drive viral uptake, often through endocytic pathways followed by trafficking through endosomes and lysosomes60, or through fusion at the plasma membrane61. Viruses must exit vesicles for productive infection, through endosomal escape or viral fusion, followed by uncoating and release of the genome, which can be cytoplasmic or nuclear. The genome is transcribed into mRNA if necessary, translated (often into large complex polyproteins that are processed by viral or host proteases) and replicated. Many emerging RNA viruses encode their own polymerases62, but some viruses also utilize host polymerases63. Viruses universally utilize host translation machinery, although viral translation is regulated in a differential manner from the cell64. Once structural proteins and genomes are created, viral particles are assembled, followed by viral egress or cell lysis65, at which point the cycle begins anew. Each of these steps in the general viral life cycle can be targeted by therapeutics.
a, Schematic of a generic viral replication cycle; different steps are illustrated in cartoon form. Text boxes indicate general steps during infection that can be targeted for antiviral therapeutics. b, Schematic of phases of human disease progression. Disease is separated into a viral phase, which can be treated by antiviral drugs, and an inflammatory phase, which can be treated by anti-inflammatory and immunomodulatory drugs. Symptomatic onset occurs during the viral phase, but the incubation time can vary.
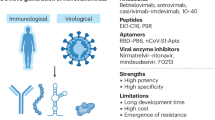
There are two fundamental types of antiviral therapeutics, direct-acting and host-factor, which differ by targeting viral or cellular factors (see Table 1 for an overview). Direct-acting antivirals target the virus itself—antibodies that bind and neutralize viral particles10, receptor decoys66,67, viral protease inhibitors68,69, nucleoside analogs4, viral translation inhibitors70 and others. Host-factor antivirals target elements of the host cell required for efficient viral infection or pathogenesis—antibodies that bind the receptor71, endocytosis inhibitors72,73, host protease inhibitors74, lipidomic reprogramming drugs75, kinase inhibitors (for example, baricitinib)9 and others. Both direct-acting and host-factor therapeutics can be broadly applicable if the antiviral targets conserved genes/motifs within a virus family (such as the coronavirus RNA-dependent RNA polymerase or 3CL protease, which have greater than 60% and 40% sequence identify, respectively)76,77, or if diverse virus family members co-opt the same host pathways to promote virus replication and/or pathogenesis (such as the furin protease)78. CRISPR screens are powerful tools to identify required host factors as putative targets79. However, host-factor therapeutics may have greater toxicity due to disruption of cellular pathways in both infected and healthy cells.
In addition to the viral life cycle, there is a defined disease progression within an organism as infection proceeds (Fig. 2b). The initial stages of disease encompass the viral phase, during which symptoms appear, with incubation times varying from about 1 to 14 days80. As disease progresses, the viral phase is replaced by an inflammatory phase, in which the body’s antiviral inflammatory responses begin to control viral replication, often damaging the body in the process81. Antiviral therapeutic treatment windows are typically limited to the viral phase of infection, after which they are ineffective82. These limited treatment windows render testing crucial in outbreak situations83. In the inflammatory phase, treatment options include anti-inflammatory and immunomodulatory drugs, such as corticosteroids, which dampen the host immune response in an attempt to control inflammatory damage84. The corticosteroid dexamethasone is currently in use for treatment of hospitalized patients with COVID-19 (ref. 5). Baricitinib, which has been approved as an emergency use COVID-19 treatment, is a kinase inhibitor in the JAK/STAT pathway, leading to lowered cytokine release85. An alternative approach is to treat earlier in disease with cytokines such as interferon-λ, which exert antiviral effects without causing tissue damage86.
RNA-based therapeutics
Types of RNA-based therapeutics
RNA-based therapeutics are a new class of therapeutics with considerable promise. In the last 20 years, multiple RNA-based therapeutics have been approved by the FDA70,87,88,89,90; many more are currently in various stages of clinical trials. These therapeutics hold great promise, as they are more modular and simpler to design than traditional drugs. RNA can act in multiple ways to exert a therapeutic effect (Fig. 3). Antisense oligonucleotides (ASOs) act through base-pairing to bind an mRNA and modulate translation, splicing or the availability of RNA-binding proteins70,91. Alternatively, anti-microRNAs (anti-miRs) directly bind microRNAs (miRNAs), blocking their ability to function92. miRNAs or short interfering RNAs (siRNAs) can also be used as therapeutics, to drive translational repression or mRNA degradation93. Small activating RNAs (saRNAs) are related to miRNAs, but are nuclear and lead to transcriptional activation94. mRNAs can be used as a therapeutic to produce protein products, or as vaccines11,12,95. RNA aptamers can be selected to bind proteins or small molecules with high affinity, in a manner similar to traditional small-molecule drugs, but with activity on a larger range of substrates96. Finally, CRISPR effectors and guide RNAs (gRNAs) can be used either to target cellular RNAs for destruction or to edit genes97. Overall, RNA-based products can be used for a variety of functions, including modulation of gene expression, splicing and translation.
Delivery of RNA-based therapeutics
A major challenge for RNA-based therapeutics is the delivery of drug to the tissue and cell type of interest. RNA has no inherent targeting abilities and must rely on localized delivery, a delivery vehicle or modifications, and much research has been carried out on this area in recent years. RNA can be directed by the attachment of a targeting moiety such as a sugar, peptide, antibody or RNA aptamer98,99. For example, an RNA aptamer developed to bind the envelope glycoprotein of the human immunodeficiency virus (HIV) can be used to target siRNAs to cells infected with HIV100. Alternatively, RNA can be packaged into lipid nanoparticles, which can then be administered systemically101. Formulations of lipid nanoparticles vary widely and are a subject of continuing research, including incorporation of targeting moieties and methods of releasing RNA at the target. Finally, ‘naked’ RNA can be administered locally to a region of interest to great effect102.
RNA therapeutics for viral outbreaks
RNA therapeutics hold great promise for preventing and treating viral infections. The use of base-pairing interactions makes RNA therapeutics simple to design, as long as the sequence of the target molecule is known. They can be designed to target conserved sequence or structural motifs, creating broadly applicable therapeutics103,104. They can also readily be adapted in response to drift in viral sequences, or to related viral strains, once an effective viral target is identified. RNA therapeutics can be either direct-acting or host-factor and can target any of the stages of the viral life cycle. An RNA could knockdown an essential viral replication factor or a required host factor, thus disrupting viral infection105,106. Alternatively, RNA products could antagonize viral downregulation of host restriction factors107. Fomivirsen was the first approved antiviral ASO therapeutic, targeting the immediate-early RNA of cytomegalovirus and blocking translation70. Multiple siRNA candidates targeting the surface antigen of the hepatitis B virus (HBV) are undergoing clinical trials for chronic HBV infection, as well as an anti-miR against miR-122 as a therapeutic for hepatitis C virus (HCV) infection. Additionally, the COVID-19 pandemic has shown the great promise of mRNA therapeutics as vaccines11,12.
Pre-emptive therapeutic development
The COVID-19 pandemic has unequivocally demonstrated the need to anticipate and prepare, rather than merely react to novel emerging pathogens. Public health strategies can be implemented immediately, including disease surveillance and modeling to identify high-risk populations. This information can be used to direct therapeutic interventions to high-risk areas and populations. However, new therapeutics and vaccines cannot be created without a lag time (Fig. 4a). As an example, newly developed antibody therapeutics for COVID-19 were not approved for use until November 2020; in contrast, remdesivir, a known broad-spectrum coronavirus antiviral3, became the first approved antiviral therapy in May 2020. This is proof of the need for proactive, rather than reactive, drug development to shorten the gap before treatments are available. Researchers should aim to have at least one drug targeting each virus family with phase 1 safety testing completed ready for rapid deployment. This is not an easy task, but it is nevertheless vital. The first step is to understand the viruses that pose the greatest risk of outbreak. We discussed some virus families of concern, but viral surveillance must be continuously carried out to identify circulating viral strains and understand changes in virus sequence and spread (see Box 1)108,109.
a, Timeline of an outbreak of a novel virus, from the time of identification to the time of control. The first response to an outbreak is the institution of public health measures, followed by antiviral drugs, followed by vaccine development. The gap to development of new antiviral drugs can be shortened by proactive drug development. b, Drug development can operate by repurposing drugs, or by creating novel drugs. Drug development pipelines integrate academic, industry and government efforts to deliver antiviral therapeutics.
With this information, the research community can then begin developing portfolios of broad-based therapeutics. There are two broad paths for therapeutic development, focusing on either repurposed drugs or novel drugs (Fig. 4b). For repurposing drugs, current FDA-approved drugs are key candidates, as they will be the fastest to be approved for a new indication in an outbreak situation (see Table 2 for example FDA-approved drugs that have been repurposed for treating viral infections)110,111,112,113. Pharmaceutical companies often have multiple well-studied drugs in their pipelines with pre-existing safety and toxicity information that are also good candidates for testing (remdesivir was previously tested for treatment of EBOV114). Libraries of approved and in-pipeline drugs should be used for high-throughput screening against a variety of viruses of interest.
In contrast, novel drug development starts with the identification of key targets based on research into required, conserved host/viral factors. Examples include viral polymerases (widely targeted by nucleoside analogs), host signaling pathways (such as JAK/STAT) or lipid biosynthesis pathways75. Once a target has been identified, therapeutics of various types (small molecule, antibody, RNA and so on) can be developed. Monoclonal antibody products typically utilize patient-derived B cells to develop antibody libraries, followed by screening for highly neutralizing antibodies10. However, before a novel pathogen emerges, there may not be human patients to provide antibodies, leaving researchers to rely on animal models. Antibodies generated from immunized animals can be humanized115, but these antibodies can still elicit unwanted immunogenic responses116. Technology that replaces the entire mouse immunoglobulin locus with the human locus has shown great promise thus far, contributing to the approved casirivimab/imdevimab COVID-19 therapeutic117,118.
Given the heterogeneity of viral strains and the inability to perfectly predict the future, it is imperative to champion broadly active therapeutics by targeting conserved factors and pathways. Regardless of type, all therapeutic candidates should be evaluated on panels of diverse viral strains both past and present, as a marker of their potential efficacy against future viral strains. A highly effective but highly specific therapeutic may not be efficacious against the next emerging virus. With effort and representative strains across the phylogenetic tree, broad-based antivirals can be identified for future epidemics.
This undertaking will require considerable effort, in the form of dedicated financial support, to encourage pre-emptive research into viral outbreak preparedness. Academic researchers drive understanding of viral and host biology leading to target identification and the beginnings of drug development. Partnerships with industry can help accelerate drug development by providing funding, development of drug libraries, quality control and manufacturing expertise. Governmental agencies lend support to academic and industry endeavors through funding and regulatory guidance. We must recognize that even more virulent zoonotic viruses than SARS-CoV-2 likely exist in nature and may emerge and distribute quickly across the globe. Consonant with the loss of life and human suffering, massive economic losses dictate a critical need for substantial investments into future preparedness.
Therapeutic regimen and delivery
Although pre-clinical research can and should be carried out in advance of an outbreak, therapeutic candidates must undergo clinical trials before approval. For novel pathogens, this may occur after an outbreak begins. The FDA has a framework for emergency use authorizations to allow unapproved products to be used in emergency situations (such as pandemics) more quickly, although evidence of safety and efficacy is still required (https://www.fda.gov/emergency-preparedness-and-response/mcm-legal-regulatory-and-policy-framework/emergency-use-authorization). The FDA also has criteria known as the Animal Rule for approval of drugs without human efficacy trials. This rule applies to drugs where field studies are not feasible and human challenge is unethical, such as treatments for anthrax poisoning and plague (Yersinia pestis). In these cases, animal efficacy studies can be used instead, although safety must still be assessed in human clinical trials (https://www.fda.gov/emergency-preparedness-and-response/mcm-regulatory-science/animal-rule-summary). This rule could possibly be used to gain approval of drugs against novel viruses before emergence.
Well-designed, double-blinded clinical trials should be carefully structured to test for preventatives as well as post-infection treatments; poorly designed trials will only lead to greater public confusion119. As discussed above, there will likely be limits to effective treatment windows in humans, and some therapeutics may be effective only as preventatives or in early stages of infection80,81,82. Pre-clinical studies in laboratory animals cannot fully inform timescales in humans, although primate studies are critical for evaluation of treatment windows. Final determination of effective treatment windows must come from clinical trials, which can be complicated by the need to estimate time of infection for patients based on time of diagnosis, contact tracing and severity of symptoms.
During an outbreak, decisions must be made to evaluate single or combination therapeutics. Trials can be designed to evaluate multiple therapeutics, or both single and combination therapeutics in the same trial, but this requires larger numbers of patients to preserve statistical power9,114,120. Analysis of combination therapeutics is more complicated, as conclusions cannot be drawn about individual therapeutics. However, in an outbreak situation, it can be beneficial to test combination treatments to reduce the time and effort needed, maximize the chance of efficacy and help combat the rise of viral resistance.
Early-stage clinical trials typically evaluate multiple doses before choosing a target dose for phase 3 trials, where optimal dosing is a balance of efficacy and toxicity. In an outbreak, manufacturing capability becomes an additional factor to consider. For a newly developed therapeutic to have an effect during an outbreak, it must be manufactured and distributed to patients, a non-trivial task. Therefore, when choosing dosing, additional consideration must be given to manufacturing capabilities and the number of doses that can be produced at different dose levels, as it may dramatically affect the number of patients that can be treated.
Although any effective drug is welcome in an outbreak, orally available drugs (or other self-administered drugs) are arguably ideal, as they provide the greatest ease of dosing, and the highest chance of patient compliance with treatment regimens, an important factor in controlling an outbreak. Additionally, self-administered drugs greatly reduce the load on healthcare workers created by the need for more difficult dosing methods, especially important in healthcare systems already overburdened owing to an outbreak, as seen with the mass COVID-19 vaccination efforts currently underway.
Conclusion: systematic efforts for drug development
History has demonstrated our capacity to integrate efforts across disciplines to develop drugs and vaccines. Similar efforts are required to protect global health from future pandemics. Pandemic preparedness for future twenty-first-century threats will be dependent on integrated research teams spanning academic, federal, industrial and philanthropic groups to identify gaps and research opportunities, coordinate interactions between teams of scientists, review progress and integrate outcomes with commercial interests. One possible model for this type of system is the antibacterial consortium CARB-X (https://carb-x.org/), although other models exist (https://www.readdi.org/). Expediting rapid pandemic responses will require pre-existing interactions with regulatory agencies to facilitate rapid therapeutic development. Commercial partners provide critical expertise in manufacturing, quality control, product testing and safety, essential to overall efforts. We recognize intellectual property as a potential impediment to product development, but this may be overcome with a flexible management scheme allowing multiple pathways to proprietary rights while retaining access for at-risk populations globally. Considering the catastrophic human and economic suffering caused by the COVID-19 pandemic, an organized global effort for pandemic preparedness represents a logical goal bridging interests across nations. The costs are minimal compared to the economic and human costs of being unprepared, especially considering that viruses capable of causing considerably higher mortality rates exist in nature. The world already saw modest outbreaks in 2003 and 2012 with the highly virulent SARS-CoV and MERS-CoV strains, and the existence of highly pathogenic influenza viruses, bunyaviruses and henipaviruses is well known. Sadly, human memory is short, so the need to act quickly is clear. We must mobilize systematic efforts to increase global preparedness against future pandemic threats, of which broadly acting antiviral therapeutics represent a critical piece. Our children’s welfare depends on us acting now.
References
Lu, R. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 (2020).
Menachery, V. D. et al. SARS-like WIV1-CoV poised for human emergence. Proc. Natl Acad. Sci. USA 113, 3048–3053 (2016).
Sheahan, T. P. et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9, eaal3653 (2017).
Sheahan, T. P. et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 12, eabb5883 (2020).
The RECOVERY Collaborative Group. Dexamethasone in hospitalized patients with Covid-19—preliminary report. N. Engl. J. Med. https://doi.org/10.1056/nejmoa2021436 (2020).
Chen, P. et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N. Engl. J. Med. https://doi.org/10.1056/nejmoa2029849 (2020).
Weinreich, D. M. et al. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N. Engl. J. Med. https://doi.org/10.1056/nejmoa2035002 (2020).
Beigel, J. H. et al. Remdesivir for the treatment of Covid-19—final report. N. Engl. J. Med. 383, 1813–1826 (2020).
Kalil, A. C. et al. Baricitinib plus remdesivir for hospitalized adults with Covid-19. N. Engl. J. Med. https://doi.org/10.1056/nejmoa2031994 (2020).
Wec, A. Z. et al. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 369, 731–736 (2020).
Baden, L. R. et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. https://doi.org/10.1056/nejmoa2035389 (2020).
Polack, F. P. et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 383, 2603–2615 (2020).
Ramasamy, M. N. et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet 396, 1979–1993 (2020).
Rappazzo, C. G. et al. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science 371, 823–829 (2021).
Menachery, V. D. et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513 (2015).
Edwards, C. E. et al. Swine acute diarrhea syndrome coronavirus replication in primary human cells reveals potential susceptibility to infection. Proc. Natl Acad. Sci. USA 117, 26915–26925 (2020).
Cherry, J. D. The chronology of the 2002–2003 SARS mini pandemic. Paediatr. Respir. Rev. 5, 262–269 (2004).
Sharif-Yakan, A. & Kanj, S. S. Emergence of MERS-CoV in the Middle East: origins, transmission, treatment, and perspectives. PLoS Pathog. 10, 10–13 (2014).
Li, W. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–680 (2005).
Luo, C.-M. et al. Discovery of novel bat coronaviruses in South China that use the same receptor as Middle East respiratory syndrome coronavirus. J. Virol. 92, e00116-18 (2018).
Gulber, D. J. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10, 100–103 (2002).
Thomas, S. J. & Yoon, I. K. A review of Dengvaxia®: development to deployment. Hum. Vaccines Immunother. 15, 2295–2314 (2019).
Kauffman, E. B. & Kramer, L. D. Zika virus mosquito vectors: competence, biology, and vector control. J. Infect. Dis. 216, S976–S990 (2017).
Glezen, W. P. Emerging infections: pandemic influenza. Epidemiol. Rev. 18, 64–76 (1996).
Osterholm, M. T. et al. Transmission of Ebola viruses: what we know and what we do not know. mBio 6, e00137 (2015).
Neumann, G., Noda, T. & Kawaoka, Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459, 931–939 (2009).
Neumann, G., Chen, H., Gao, G. F., Shu, Y. & Kawaoka, Y. H5N1 influenza viruses: outbreaks and biological properties. Cell Res. 20, 51–61 (2010).
Nachbagauer, R. et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 27, 106–114 (2021).
Holmes, E. C., Dudas, G., Rambaut, A. & Andersen, K. G. The evolution of Ebola virus: insights from the 2013–2016 epidemic. Nature 538, 193–200 (2016).
Ilunga Kalenga, O. et al. The ongoing Ebola epidemic in the Democratic Republic of Congo, 2018–2019. N. Engl. J. Med. 381, 373–383 (2019).
Henao-Restrepo, A. M. et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!). Lancet 389, 505–518 (2017).
Arankalle, V. A. et al. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J. Gen. Virol. 88, 1967–1976 (2007).
Burt, F. J. et al. Chikungunya virus: an update on the biology and pathogenesis of this emerging pathogen. Lancet Infect. Dis. 17, e107–e117 (2017).
Lindsey, N. P., Staples, J. E. & Fischer, M. Eastern equine encephalitis virus in the United States, 2003–2016. Am. J. Trop. Med. Hyg. 98, 1472–1477 (2018).
Forrester, N. L. et al. Evolution and spread of Venezuelan equine encephalitis complex alphavirus in the Americas. PLoS Negl. Trop. Dis. 11, 1–19 (2017).
Bente, D. A. et al. Crimean-Congo hemorrhagic fever: history, epidemiology, pathogenesis, clinical syndrome and genetic diversity. Antivir. Res. 100, 159–189 (2013).
Jonsson, C. B., Figueiredo, L. T. M. & Vapalahti, O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin. Microbiol. Rev. 23, 412–441 (2010).
Garnett, L. E. & Strong, J. E. Lassa fever: with 50 years of study, hundreds of thousands of patients and an extremely high disease burden, what have we learned? Curr. Opin. Virol. 37, 123–131 (2019).
Himeidan, Y. E., Kweka, E. J., Mahgoub, M. M., El Rayah, E. A. & Ouma, J. O. Recent outbreaks of Rift Valley fever in East Africa and the Middle East. Front. Public Health https://doi.org/10.3389/fpubh.2014.00169 (2014).
Pillai, V. S., Krishna, G. & Veettil, M. V. Nipah virus: past outbreaks and future containment. Viruses 12, https://doi.org/10.3390/v12040465 (2020).
Field, H. E. Hendra virus ecology and transmission. Curr. Opin. Virol. 16, 120–125 (2016).
Middleton, D. et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis. 20, 372–379 (2014).
Chhabra, P. et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 100, 1393–1406 (2019).
De Graaf, M., Van Beek, J. & Koopmans, M. P. G. Human norovirus transmission and evolution in a changing world. Nat. Rev. Microbiol. 14, 421–433 (2016).
Epstein, J. H. & Anthony, S. J. Viral discovery as a fool for pandemic preparedness. Rev. Sci. Tech. 36, 499–512 (2017).
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382, 727–733 (2020).
Messer, W. B. et al. Development and characterization of a reverse genetic system for studying dengue virus serotype 3 strain variation and neutralization. PLoS Negl. Trop. Dis. 6, e1486 (2012).
Yount, B. et al. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl Acad. Sci. USA 100, 12995–13000 (2003).
Hou, Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 1468, eabe8499 (2020).
Kim, J. II et al. GFP-expressing influenza a virus for evaluation of the efficacy of antiviral agents. J. Microbiol. 50, 359–362 (2012).
Zou, G., Xu, H. Y., Qing, M., Wang, Q. Y. & Shi, P. Y. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antivir. Res. 91, 11–19 (2011).
Dijkman, R. et al. Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 87, 6081–6090 (2013).
Tang, H., Abouleila, Y. & Mashaghi, A. Lassa hemorrhagic shock syndrome-on-a-chip. Biotechnol. Bioeng. https://doi.org/10.1002/bit.27636 (2020).
Zhang, M. et al. Biomimetic human disease model of SARS-CoV-2-induced lung injury and immune responses on organ chip system. Adv. Sci. https://doi.org/10.1002/advs.202002928 (2020).
Yang, L. et al. A human pluripotent stem cell-based platform to study SARS-CoV-2 tropism and model virus infection in human cells and organoids. Cell Stem Cell 27, 125–136 (2020).
The Complex Trait Consortium. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 36, 1133–1137 (2018)..
Sun, J. et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182, 734–743 (2020).
Sun, S. H. et al. A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28, 124–133 (2020).
Li, W. et al. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Nature 426, 450–454 (2003).
Milewska, A. et al. Entry of human coronavirus NL63 into the cell. J. Virol. 92, e01933-17 (2018).
Millet, J. K. & Whittaker, G. R. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology 517, 3–8 (2018).
Kirchdoerfer, R. N. & Ward, A. B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 10, 2342 (2019).
De Villiers, J., Schaffner, W., Tyndall, C., Lupton, S. & Kamen, R. Polyoma virus DNA replication requires an enhancer. Nature 312, 242–246 (1984).
Jan, E., Mohr, I. & Walsh, D. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Annu. Rev. Virol. 3, 283–307 (2016).
Neuman, B. W. et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 174, 11–22 (2011).
Glasgow, A. et al. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc. Natl Acad. Sci. USA 117, 28046–28055 (2020).
Linsky, T. W. et al. De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science 370, 1208–1214 (2020).
Klemm, T. et al. Mechanism and inhibition of the papain‐like protease, PLpro, of SARS‐CoV‐2. EMBO J. 39, e106275 (2020).
Chen, C.-C. et al. Overview of antiviral drug candidates targeting coronaviral 3C-like main proteases. FEBS J. https://doi.org/10.1111/febs.15696 (2021).
Anderson, K. P., Fox, M. C., Brown-Driver, V., Martin, M. J. & Azad, R. F. Inhibition of human cytomegalovirus immediate-early gene expression by an antisense oligonucleotide complementary to immediate-early RNA. Antimicrob. Agents Chemother. 40, 2004–2011 (1996).
Raj, V. S. et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495, 251–254 (2013).
Ivanov, A. I. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol. Biol. 440, 15–33 (2008).
Inoue, Y. et al. Clathrin-dependent entry of severe acute respiratory syndrome coronavirus into target cells expressing ACE2 with the cytoplasmic tail deleted. J. Virol. 81, 8722–8729 (2007).
Zhou, Y. et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 116, 76–84 (2015).
Yuan, S. et al. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun. 10, 120 (2019).
Zhang, W. F. et al. Novel coronavirus polymerase and nucleotidyl-transferase structures: potential to target new outbreaks. J. Phys. Chem. Lett. 11, 4430–4435 (2020).
Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R. & Hilgenfeld, R. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300, 1763–1767 (2003).
Braun, E. & Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 8, 1–19 (2019).
Wei, J. et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell 184, 76–91 (2021).
Linton, N. et al. Incubation period and other epidemiological characteristics of 2019 novel coronavirus infections with right truncation: a statistical analysis of publicly available case data. J. Clin. Med. 9, 538 (2020).
Peiris, J. S. M. et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361, 1767–1772 (2003).
Widagdo, W., Okba, N. M. A., Stalin Raj, V. & Haagmans, B. L. MERS-coronavirus: from discovery to intervention. One Health 3, 11–16 (2017).
Ahmed, A. E. Diagnostic delays in Middle East respiratory syndrome coronavirus patients and health systems. J. Infect. Public Health 12, 767–771 (2019).
Yang, J. W., Yang, L., Luo, R. G. & Xu, J. F. Corticosteroid administration for viral pneumonia: COVID-19 and beyond. Clin. Microbiol. Infect. 26, 1171–1177 (2020).
Stebbing, J. et al. Mechanism of baricitinib supports artificial intelligence‐predicted testing in COVID‐19 patients. EMBO Mol. Med. 12, e12697 (2020).
Andreakos, E. & Tsiodras, S. COVID‐19: lambda interferon against viral load and hyperinflammation. EMBO Mol. Med. 12, 10–13 (2020).
Ng, E. W. M. et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 5, 123–132 (2006).
Coelho, T. et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 369, 819–829 (2013).
Benson, M. D. et al. Inotersen treatment for patients with hereditary transthyretin amyloidosis. N. Engl. J. Med. 379, 22–31 (2018).
Balwani, M. et al. Phase 3 trial of RNAi therapeutic givosiran for acute intermittent porphyria. N. Engl. J. Med. 382, 2289–2301 (2020).
Hua, Y. et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 24, 1634–1644 (2010).
Lanford, R. E. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327, 198–201 (2010).
Giladi, H. et al. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther. 8, 769–776 (2003).
Sarker, D. et al. MTL-CEBPA, a small activating RNA therapeutic upregulating C/EBP-α, in patients with advanced liver cancer: a first-in-human, multicenter, open-label, phase I trial. Clin. Cancer Res. 26, 3936–3946 (2020).
Kose, N. et al. A lipid-encapsulated mRNA encoding a potently neutralizing human monoclonal antibody protects against chikungunya infection. Sci. Immunol. 4, eaaw6647 (2019).
Jellinek, D., Green, L. S., Bell, C. & Janjić, N. Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry 33, 10450–10456 (1994).
Abbott, T. R. et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 181, 865–876 (2020).
Prakash, T. P. et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 42, 8796–8807 (2014).
Sugo, T. et al. Development of antibody-siRNA conjugate targeted to cardiac and skeletal muscles. J. Control. Release 237, 1–13 (2016).
Zhou, J. et al. Receptor-targeted aptamer-siRNA conjugate-directed transcriptional regulation of HIV-1. Theranostics 8, 1575–1590 (2018).
Veiga, N. et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 9, 4493 (2018).
Carlsson, L. et al. Biocompatible, purified VEGF-A mRNA improves cardiac function after intracardiac injection 1 week post-myocardial infarction in swine. Mol. Ther. Methods Clin. Dev. 9, 330–346 (2018).
Ahn, D. G. et al. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antivir. Res. 91, 1–10 (2011).
Szabat, M. et al. RNA secondary structure as a first step for rational design of the oligonucleotides towards inhibition of influenza a virus replication. Pathogens 9, 925 (2020).
Whitten-Bauer, C. et al. The host factor Erlin-1 is required for efficient hepatitis C virus infection. Cells 8, 1555 (2019).
Meng, B., Lui, Y. W., Meng, S., Cao, C. & Hu, Y. Identification of effective siRNA blocking the expression of SARS viral envelope E and RDRP genes. Mol. Biotechnol. 33, 141–148 (2006).
Gorbea, C., Mosbruger, T. & Cazalla, D. A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis. Nature 550, 275–279 (2017).
Flies, E. J., Toi, C., Weinstein, P., Doggett, S. L. & Williams, C. R. Converting mosquito surveillance to arbovirus surveillance with honey-baited nucleic acid preservation cards. Vector-Borne Zoonotic Dis. 15, 397–403 (2015).
Phelps, K. L. et al. Bat research networks and viral surveillance: gaps and opportunities in western Asia. Viruses 11, 240 (2019).
Yang, L. et al. Identification of SARS-CoV-2 entry inhibitors among already approved drugs. Acta Pharmacol. Sin. https://doi.org/10.1038/s41401-020-00556-6 (2020).
Riva, L. et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 586, 113–119 (2020).
De Wilde, A. H. et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 58, 4875–4884 (2014).
Tan, E. L. C. et al. Inhibition of SARS coronavirus infection in vitro with clinically approved antiviral drugs. Emerg. Infect. Dis. 10, 581–586 (2004).
Mulangu, S. et al. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 381, 2293–2303 (2019).
Li, Y. et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 25, 1237–1249 (2015).
Harding, F. A., Stickler, M. M., Razo, J. & DuBridge, R. B. The immunogenicity of humanized and fully human antibodies: residual immunogenicity resides in the CDR regions. MAbs 2, 256–265 (2010).
Pascal, K. E. et al. Pre- and postexposure efficacy of fully human antibodies against spike protein in a novel humanized mouse model of MERS-CoV infection. Proc. Natl Acad. Sci. USA 112, 8738–8743 (2015).
Hansen, J. et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020).
Hashem, H., Abufaraj, M., Tbakhi, A., & Sultan, I. Obstacles and considerations related to clinical trial research during the COVID-19 pandemic. Front. Med. 7, 598038 (2020).
Hung, I. F. N. et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 395, 1695–1704 (2020).
Haffizulla, J. et al. Effect of nitazoxanide in adults and adolescents with acute uncomplicated influenza: a double-blind, randomised, placebo-controlled, phase 2b/3 trial. Lancet Infect. Dis. 14, 609–618 (2014).
Rossignol, J. F., Abu-Zekry, M., Hussein, A. & Santoro, M. G. Effect of nitazoxanide for treatment of severe rotavirus diarrhoea: randomised double-blind placebo-controlled trial. Lancet 368, 124–129 (2006).
Cai, Q. et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering 6, 1192–1198 (2020).
Sissoko, D. et al. Experimental treatment with favipiravir for Ebola virus disease (the JIKI trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med. 13, e1001967 (2016).
Eberhardt, K. A. et al. Ribavirin for the treatment of Lassa fever: a systematic review and meta-analysis. Int. J. Infect. Dis. 87, 15–20 (2019).
Chong, H. T. et al. Treatment of acute Nipah encephalitis with ribavirin. Ann. Neurol. 49, 810–813 (2001).
Hung, I. F. N. et al. Efficacy of clarithromycin-naproxen-oseltamivir combination in the treatment of patients hospitalized for influenza A(H3N2) infection: an open-label randomized, controlled, phase IIb/III trial. Chest 151, 1069–1080 (2017).
Hampson, L. et al. A single-arm, proof-of-concept trial of Lopimune (lopinavir/ritonavir) as a treatment for HPV-related pre-invasive cervical disease. PLoS ONE 11, e0147917 (2016).
Acknowledgements
We thank L. P. V. Tse for critical reading of this manuscript and helpful discussions. The manuscript presented here was supported by grants AI108197, AI132178, AI42759 and AI157155 from the National Institute of Allergy and Infectious Disease of the United States NIH. This project was also supported by the North Carolina Policy Collaboratory at the University of North Carolina at Chapel Hill with funding from the North Carolina Coronavirus Relief Fund established and appropriated by the North Carolina General Assembly.
Author information
Authors and Affiliations
Contributions
R.M.M. wrote the review. R.S.B. reviewed and revised the final version.
Corresponding author
Ethics declarations
Competing interests
R.S.B. has competing interests regarding collaborative research with Gilead Sciences, Ridgeback Biotherapeutics, Takeda, Pfizer, Eli Lily, Adimab and Moderna Therapeutics.
Additional information
Peer review information Nature Medicine thanks Pei-Yong Shi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Joao Monteiro was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Meganck, R.M., Baric, R.S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat Med 27, 401–410 (2021). https://doi.org/10.1038/s41591-021-01282-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-021-01282-0
This article is cited by
-
Therapeutic strategies for COVID-19: progress and lessons learned
Nature Reviews Drug Discovery (2023)
-
Pharmacological disruption of mSWI/SNF complex activity restricts SARS-CoV-2 infection
Nature Genetics (2023)
-
Cas13d knockdown of lung protease Ctsl prevents and treats SARS-CoV-2 infection
Nature Chemical Biology (2022)
-
2′–5′ oligoadenylate synthetase‑like 1 (OASL1) protects against atherosclerosis by maintaining endothelial nitric oxide synthase mRNA stability
Nature Communications (2022)
-
Effects of Boswellia species on viral infections with particular attention to SARS-CoV-2
Inflammopharmacology (2022)