Abstract
Proteins are effector molecules that mediate the functions of genes1,2 and modulate comorbidities3,4,5,6,7,8,9,10, behaviors and drug treatments11. They represent an enormous potential resource for personalized, systemic and data-driven diagnosis, prevention, monitoring and treatment. However, the concept of using plasma proteins for individualized health assessment across many health conditions simultaneously has not been tested. Here, we show that plasma protein expression patterns strongly encode for multiple different health states, future disease risks and lifestyle behaviors. We developed and validated protein-phenotype models for 11 different health indicators: liver fat, kidney filtration, percentage body fat, visceral fat mass, lean body mass, cardiopulmonary fitness, physical activity, alcohol consumption, cigarette smoking, diabetes risk and primary cardiovascular event risk. The analyses were prospectively planned, documented and executed at scale on archived samples and clinical data, with a total of ~85 million protein measurements in 16,894 participants. Our proof-of-concept study demonstrates that protein expression patterns reliably encode for many different health issues, and that large-scale protein scanning12,13,14,15,16 coupled with machine learning is viable for the development and future simultaneous delivery of multiple measures of health. We anticipate that, with further validation and the addition of more protein-phenotype models, this approach could enable a single-source, individualized so-called liquid health check.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
Pre-existing data access policies for each of the five parent cohort studies specify that research data requests can be submitted to each steering committee; these will be promptly reviewed for confidentiality or intellectual property restrictions and will not unreasonably be refused. Individual-level patient or protein data may further be restricted by consent, confidentiality or privacy laws/considerations. These policies apply to both clinical and proteomic data.
References
Sun, B. B. et al. Genomic atlas of the human plasma proteome. Nature 558, 73–79 (2018).
Emilsson, V. et al. Co-regulatory networks of human serum proteins link genetics to disease. Science 361, 769–773. (2018).
Tasaki, S. et al. Multi-omics monitoring of drug response in rheumatoid arthritis in pursuit of molecular remission. Nat. Commun. 9, 2755 (2018).
O’Dwyer, D. N. et al. The peripheral blood proteome signature of idiopathic pulmonary fibrosis is distinct from normal and is associated with novel immunological processes. Sci. Rep. 7, 46560 (2017).
Christensson, A. et al. The impact of the glomerular filtration rate on the human plasma proteome. Proteom. Clin. Appl. 12, e1700067 (2018).
Ganz, P. et al. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. J. Am. Med. Assoc. 315, 2532–2541 (2016).
Wood, G. C., Chu, X. & Argyropoulos, G. et al. A multi-component classifier for nonalcoholic fatty liver disease (NAFLD) based on genomic, proteomic, and phenomic data domains. Sci. Rep. 7, 43238 (2017).
Han, Z. et al. Validation of a novel modified aptamer-based array proteomic platform in patients with end-stage renal disease. Diagnostics (Basel) 8, 71 (2018).
Menni, C. et al. Circulating proteomic signatures of chronolological age. J. Gerontol. A 70, 809–816 (2014).
Thrush, A. et al. Diet-resistant obesity is characterized by a distinct plasma proteomic signature and impaired muscle fiber metabolism. Int. J. Obes. 42, 353–362 (2018).
Williams, S. A. et al. Improving assessment of drug safety through proteomics: early detection and mechanistic characterization of the unforeseen harmful effects of torcetrapib. Circulation 137, 999–1010 (2018).
Rohloff, J. C. et al. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol. Ther. Nucleic Acids 3, e201 (2014).
Gold, L. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE 5, e15004 (2010).
Brody, E. et al. Life’s simple measures: unlocking the proteome. J. Mol. Biol. 422, 595–606 (2012).
Kim, C. H. et al. Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci. Rep. 8, 8382 (2018).
Candia, J. et al. Assessment of variability in the SOMAscan assay. Sci. Rep. 7, 14248 (2017).
Collaborators GBDRF, Forouzanfar, M. H. et al. Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 386, 2287–2323 (2015).
Maruthappu, M. Delivering triple prevention: a health system responsibility. Lancet Diabetes Endocrinol. 4, 299–301 (2016).
Robson, J. et al. The NHS Health Check in England: an evaluation of the first 4 years. BMJ Open 6, e008840 (2016).
Valabhji, J. et al. Efficacy and effectiveness of screen and treat policies in prevention of type 2 diabetes: systematic review and meta-analysis of screening tests and interventions. BMJ 356, i6538 (2017).
Middleton, K. R., Anton, S. D. & Perri, M. G. Long-term adherence to health behavior change. Am. J. Lifestyle Med. 7, 395–404 (2013).
Dimitrov, D. V. Medical internet of things and big data in healthcare. Health Inf. Res. 22, 156–163 (2016).
Flores, M., Glusman, G., Brogaard, K., Price, N. D. & Hood, L. P4 medicine: how systems medicine will transform the healthcare sector and society. Per. Med. 10, 565–576 (2013).
Musich, S., Wang, S., Hawkins, K. & Klemes, A. The impact of personalized preventive care on health care quality, utilization, and expenditures. Popul. Health Manag. 19, 389–397. (2016).
Ezkurdia, I. et al. Multiple evidence strands suggest that there may be as few as 19,000 human protein-coding genes. Hum. Mol. Genet. 23, 5866–5878 (2014).
Lin, H. et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science 320, 807–811 (2008).
Harrell, F. E. Jr. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis (Springer, 2015).
Pencina, Michael J. et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat. Med. 27, 157–172 (2008).
Fielding, C. M. & Angulo, P. Hepatic steatosis and steatohepatitis: are they really two distinct entities? Curr. Hepatol. Rep. 13, 151–158 (2014).
Yki-Jarvinen, H. Non-alcoholic fatty liver disease as a cause and a consequence of metabolic syndrome. Lancet Diabetes Endocrinol. 2, 901–910. (2014).
Shuster, A., Patlas, M., Pinthus, J. H. & Mourtzakis, M. The clinical importance of visceral adiposity: a critical review of methods for visceral adipose tissue analysis. Br. J. Radiol. 85, 1–10 (2012).
Ross, R. et al. Importance of assessing cardiorespiratory fitness in clinical practice: a case for fitness as a clinical vital sign: a scientific statement from the American Heart Association. Circulation 134, e653–e699 (2016).
de Souza de Silva, C. G. et al. Association between cardiorespiratory fitness, obesity, and health care costs: The Veterans Exercise Testing Study. Int. J. Obes. (Lond.) https://doi.org/10.1038/s41366-018-0257-0 (2018).
Hobbs, F. D., Jukema, J. W., Da Silva, P. M., McCormack, T. & Catapano, A. L. Barriers to cardiovascular disease risk scoring and primary prevention in Europe. QJM 103, 727–739 (2010).
Ostroff, R. M. et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS ONE 5, e15003 (2010).
Ostroff, R. M. et al. Early detection of malignant pleural mesothelioma in asbestos-exposed individuals with a noninvasive proteomics-based surveillance tool. PLoS ONE 7, e46091 (2012).
Usher-Smith, J. A., Sharp, S. J. & Griffin, S. J. The spectrum effect in tests for risk prediction, screening, and diagnosis. BMJ 353, i3139 (2016).
Ganna, A. et al. Risk prediction measures for case-cohort and nested case-control designs: an application to cardiovascular disease. Am. J. Epidemiol. 175, 715–724 (2012).
Levey, A. S. et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 150, 604–612 (2009).
Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 67, 301–320 (2005).
Tibshirani, R. Regression shrinkage and selection via the LASSO. J. R. Stat. Soc. B 58, 267–288 (1996).
Acknowledgements
The Whitehall II study is supported by the UK Medical Research Council UK (no. MR/R024227/1, to M.K.), the US National Institutes on Aging (NIH, nos. US R01AG056477, R01AG062553) to M.K. and the British Heart Foundation (no. RG/16/11/32334) to M.J.S. A.D.H. is a NIHR Senior Investigator and was also supported, in part, by the National Institute for Health Research University College London Hospitals Biomedical Research Centre and the UCL BHF Research Accelerator (AA/18/6/34223). FENLAND (the Fenland study, no. 10.22025/2017.10.101.00001) is funded by UK Medical Research Council (no. MC_UU_12015/1), and N.W. is a NIHR senior investigator. We also thank the Fenland Study Investigators, Fenland Study Co-ordination team and the Epidemiology Field, Data and Laboratory teams. HUNT3 is funded by the Norwegian Ministry of Health, Norwegian University of Science and Technology and Norwegian Research Council, Central Norway Regional Health Authority, the Nord-Trondelag County Council and the Norwegian Institute of Public Health. The HERITAGE Family study was funded by the US National Heart, Lung and Blood Institute grants (NIH/NHLBI, no. R01HL146462 to M.A.S.) and no. HL45670 (HERITAGE, to C.B.). All authors are grateful to all volunteers/participants in all of the cohorts, and to the general practitioners, other physicans and practice staff for assistance with recruitment. SomaScan assays and the Covance study were funded by SomaLogic, Inc. The authors also thank A. Lowell (leader of the SomaLogic assay team), D. Perry for the bioinformatics of quality control, J. Williams for the agreements with the study institutions and J. Zach for clinical data organization and management.
Author information
Authors and Affiliations
Contributions
In an academic–industry partnership, SomaLogic, Inc. and the academic collaborators worked together on study design, interpretation of the data and preparation of the manuscript. S.A.W., P.G. and N.W. were responsible for designing, writing and final editing of the manuscript and responses to reviewer comments. In addition to all authors being generally involved in the program, specific contributions were as follows: M.K. and M.J.S. were accountable for the data from the Whitehall II study and advised on the study design for the CV and diabetes models. C.L. and N.W. were accountable for the data from the Fenland study and advising on diabetes risk and behavioral models. C.B. and M.A.S. were accountable for the data from the Heritage Family study. C.J. was accountable for the data from the HUNT3 study. R.O. was accountable for the data from the Covance study. L.A., G.D., R.K.D., Y.H., M.H. and S.W. designed and executed the machine learning tactics and developed the models. R.O., J.A., T.B., J.C. and S.A.W. were responsible for the design and integration of the program across studies. A.D.H. and J.P.C. were particularly involved in the design, execution and interpretation of the CV risk evaluations.
Corresponding author
Ethics declarations
Competing interests
The SomaLogic co-authors (S.W., L.A., J.A., T.B., J.C., G.D., R.K.D., Y.H., M.H., R.O. and S.W.) were/are all employees of SomaLogic, Inc., which has a commercial interest in the results. N.W. and C.L. declared that SomaLogic, Inc. has given a grant to the University of Cambridge. P.G. is a member of the SomaLogic Medical Advisory board, for which he receives no remuneration of any kind. The remaining authors (M.K., A.H., J.P.C., C.B., C.J., M.S. and M.S.) have no competing interests.
Additional information
Peer review information Jennifer Sargent was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
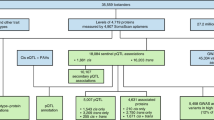
Extended Data Fig. 1 Descriptors of parent studies and fractions used for model derivation and validation.
Solid black arrows designate how fractions of samples and clinical data were utilized independently; blue dashed arrows designate the validation of finalized models either in new fractions of the same dataset or in independent datasets. eGFR = estimated glomerular filtration rate; VO2max. = maximum rate of oxygen consumption; kg. = kilograms. *For Fenland, the precise numbers available for 70%/15%/15% fractions depended on the numbers of participants with data for each endpoint as follows: n=9654 for self-reported alcohol units, n = 11,471 with DEXA scans for body composition, n=10,077 with ultrasound for liver fat, n=11,695 with individually calibrated heart rate and movement sensing for caloric expenditure due to physical activity. **For HERITAGE the model was trained on the pre-training time point from half the 523 participants and the post training time point from the other half of the participants. The model was tested on samples with the opposite time points in the same participants and finally replicated in the 10% fraction not used for training.
Extended Data Fig. 2
Details of the 5 parent cohort studies.
Extended Data Fig. 3
Participant characteristics for current health state models.
Extended Data Fig. 4
Participant characteristics for current state body composition models.
Extended Data Fig. 5
Participant characteristics for modifiable behavioral factors models.
Extended Data Fig. 6
Participant characteristics for future metabolic health risks models.
Supplementary information
Supplementary Tables
Supplementary Tables 1–6.
Source data
Source Data Fig. 1
Statistical Source Data for 12 individual panels in Fig. 1
Rights and permissions
About this article
Cite this article
Williams, S.A., Kivimaki, M., Langenberg, C. et al. Plasma protein patterns as comprehensive indicators of health. Nat Med 25, 1851–1857 (2019). https://doi.org/10.1038/s41591-019-0665-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0665-2
This article is cited by
-
Adopting artificial intelligence in cardiovascular medicine: a scoping review
Hypertension Research (2024)
-
Biological aging as a predictor of cardiometabolic multimorbidity
Nature Cardiovascular Research (2024)
-
The transition from genomics to phenomics in personalized population health
Nature Reviews Genetics (2024)
-
Multi-omic prediction of incident type 2 diabetes
Diabetologia (2024)
-
Comprehensive proteomics of CSF, plasma, and urine identify DDC and other biomarkers of early Parkinson’s disease
Acta Neuropathologica (2024)