Abstract
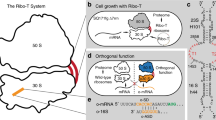
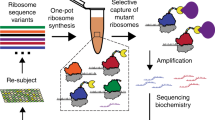
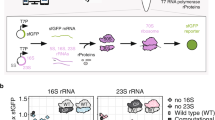
RNA-based macromolecular machines, such as the ribosome, have functional parts reliant on structural interactions spanning sequence-distant regions. These features limit evolutionary exploration of mutant libraries and confound three-dimensional structure-guided design. To address these challenges, we describe Evolink (evolution and linkage), a method that enables high-throughput evolution of sequence-distant regions in large macromolecular machines, and library design guided by computational RNA modeling to enable exploration of structurally stable designs. Using Evolink, we evolved a tethered ribosome with a 58% increased activity in orthogonal protein translation and a 97% improvement in doubling times in SQ171 cells compared to a previously developed tethered ribosome, and reveal new permissible sequences in a pair of ribosomal helices with previously explored biological function. The Evolink approach may enable enhanced engineering of macromolecular machines for new and improved functions for synthetic biology.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The authors declare that all experimental data supporting the findings of this study are available within the paper and its supplementary files. Publicly available data, such as the 4YBB (PDB) ribosome structure are mentioned explicitly when used. All data related to models are available upon request from the authors. The map and fitted model for the cryo-EM data are reported as Electron Microscopy Data Bank entry no. EMD-26666 and PDB structure 7UPH. Source data are provided with this paper.
Code availability
All inputs and command files used in setting up computational modeling are available at https://github.com/everyday847/ribotv3_simulations.
References
Dedkova, L. M., Fahmi, N. E., Golovine, S. Y. & Hecht, S. M. Enhanced d-amino acid incorporation into protein by modified ribosomes. J. Am. Chem. Soc. 125, 6616–6617 (2003).
Dedkova, L. M., Fahmi, N. E., Golovine, S. Y. & Hecht, S. M. Construction of modified ribosomes for incorporation of d-amino acids into proteins. Biochemistry 45, 15541–15551 (2006).
Dedkova, L. M. et al. β-Puromycin selection of modified ribosomes for in vitro incorporation of β-amino acids. Biochemistry 51, 401–415 (2012).
Dedkova, L. M. & Hecht, S. M. Expanding the scope of protein synthesis using modified ribosomes. J. Am. Chem. Soc. 141, 6430–6447 (2019).
Des Soye, B. J., Patel, J. R., Isaacs, F. J. & Jewett, M. C. Repurposing the translation apparatus for synthetic biology. Curr. Opin. Chem. Biol. 28, 83–90 (2015).
Ellefson, J. W. et al. Synthetic evolutionary origin of a proofreading reverse transcriptase. Science 352, 1590–1593 (2016).
Hammerling, M. J., Fritz, B. R., Yoesep, D. J., Carlson, E. D. & Jewett, M. C. In vitro ribosome synthesis and evolution through ribosome display. Nat. Commun. 11, 1108 (2020).
Maini, R. et al. Protein synthesis with ribosomes selected for the incorporation of β-amino acids. Biochemistry 54, 3694–3706 (2015).
Carlson, E. D. et al. Engineered ribosomes with tethered subunits for expanding biological function. Nat. Commun. 10, 3920 (2019).
Romero, P. A. & Arnold, F. H. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 10, 866–876 (2009).
Sailer, Z. R. & Harms, M. J. Molecular ensembles make evolution unpredictable. Proc. Natl Acad. Sci. USA 114, 11938–11943 (2017).
Sarkisyan, K. S. et al. Local fitness landscape of the green fluorescent protein. Nature 533, 397–401 (2016).
Ramakrishnan, V. Ribosome structure and the mechanism of translation. Cell 108, 557–572 (2002).
Schmied, W. H. et al. Controlling orthogonal ribosome subunit interactions enables evolution of new function. Nature 564, 444–448 (2018).
Fried, S. D., Schmied, W. H., Uttamapinant, C. & Chin, J. W. Ribosome subunit stapling for orthogonal translation in E. coli. Angew. Chem. 127, 12982–12985 (2015).
Liu, C. C., Jewett, M. C., Chin, J. W. & Voigt, C. A. Toward an orthogonal central dogma. Nat. Chem. Biol. 14, 103–106 (2018).
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M. & Chin, J. W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444 (2010).
Orelle, C. et al. Protein synthesis by ribosomes with tethered subunits. Nature 524, 119–124 (2015).
Rackham, O. & Chin, J. W. A network of orthogonal ribosome· mRNA pairs. Nat. Chem. Biol. 1, 159–166 (2005).
Wang, K., Neumann, H., Peak-Chew, S. Y. & Chin, J. W. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25, 770–777 (2007).
Liu, Y., Kim, D. S. & Jewett, M. C. Repurposing ribosomes for synthetic biology. Curr. Opin. Chem. Biol. 40, 87–94 (2017).
Liu, F., Bratulić, S., Costello, A., Miettinen, T. P. & Badran, A. H. Directed evolution of rRNA improves translation kinetics and recombinant protein yield. Nat. Commun. 12, 5638 (2021).
Goto, Y., Katoh, T. & Suga, H. Flexizymes for genetic code reprogramming. Nat. Protoc. 6, 779–790 (2011).
Melo Czekster, C., Robertson, W. E., Walker, A. S., Söll, D. & Schepartz, A. In vivo biosynthesis of a β-amino acid-containing protein. J. Am. Chem. Soc. 138, 5194–5197 (2016).
Chin, J. W. Expanding and reprogramming the genetic code. Nature 550, 53–60 (2017).
Jewett, M. C., Fritz, B. R., Timmerman, L. E. & Church, G. M. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol. Syst. Biol. 9, 678 (2013).
Hui, A. & de Boer, H. A. Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl Acad. Sci. USA 84, 4762–4766 (1987).
Rackham, O. & Chin, J. W. Cellular logic with orthogonal ribosomes. J. Am. Chem. Soc. 127, 17584–17585 (2005).
Cho, N. et al. De novo assembly and next-generation sequencing to analyse full-length gene variants from codon-barcoded libraries. Nat. Commun. 6, 8351 (2015).
Yoo, J. I., Daugherty, P. S. & O’Malley, M. A. Bridging non-overlapping reads illuminates high-order epistasis between distal protein sites in a GPCR. Nat. Commun. 11, 690 (2020).
Borgström, E. et al. Phasing of single DNA molecules by massively parallel barcoding. Nat. Commun. 6, 7173 (2015).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Asai, T., Zaporojets, D., Squires, C. & Squires, C. L. An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc. Natl Acad. Sci. USA 96, 1971–1976 (1999).
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Watkins, A. M., Rangan, R. & Das, R. FARFAR2: improved de novo Rosetta prediction of complex global RNA folds. Structure 28, 963–976 (2020).
Noeske, J. et al. High-resolution structure of the Escherichia coli ribosome. Nat. Struct. Mol. Biol. 22, 336–341 (2015).
Zubradt, M. et al. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat. Methods 14, 75–82 (2017).
Aleksashin, N. A. et al. Assembly and functionality of the ribosome with tethered subunits. Nat. Commun. 10, 930 (2019).
Lee, J., Schwarz, K. J., Kim, D. S., Moore, J. S. & Jewett, M. C. Ribosome-mediated polymerization of long chain carbon and cyclic amino acids into peptides in vitro. Nat. Commun. 11, 4304 (2020).
Lee, J. et al. Expanding the limits of the second genetic code with ribozymes. Nat. Commun. 10, 5097 (2019).
Lee, J., Torres, R., Byrom, M., Ellington, A. D. & Jewett, M. C. Ribosomal incorporation of cyclic β-amino acids into peptides using in vitro translation. Chem. Commun. 56, 5597–5600 (2020).
Kappel, K. et al. De novo computational RNA modeling into cryo-EM maps of large ribonucleoprotein complexes. Nat. Methods 15, 947–954 (2018).
Sun, Q., Vila-Sanjurjo, A. & O’Connor, M. Mutations in the intersubunit bridge regions of 16S rRNA affect decoding and subunit–subunit interactions on the 70S ribosome. Nucleic Acids Res. 39, 3321–3330 (2011).
Zhang, L. et al. The structural basis for inhibition of ribosomal translocation by viomycin. Proc. Natl Acad. Sci. USA 117, 10271–10277 (2020).
Pulk, A., Maiväli, Ü. & Remme, J. Identification of nucleotides in E. coli 16S rRNA essential for ribosome subunit association. RNA 12, 790–796 (2006).
Aleksashin, N. A. et al. A fully orthogonal system for protein synthesis in bacterial cells. Nat. Commun. 11, 1858 (2020).
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G. & Neufeld, J. D. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 13, 31 (2012).
Incarnato, D., Morandi, E., Simon, L. M. & Oliviero, S. RNA Framework: an all-in-one toolkit for the analysis of RNA structures and post-transcriptional modifications. Nucleic Acids Res. 46, e97–e97 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pintilie, G. D., Zhang, J., Goddard, T. D., Chiu, W. & Gossard, D. C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438 (2010).
Acknowledgements
This work was supported by the National Science Foundation (grant no. MCB-1716766), the Human Frontiers Science Program (grant no. RGP0015/2017), the Army Research Office (grant no. W911NF-16-1-0372), all to M.C.J. R.D. thanks the NIGMS MIRA R35 award for funding. We thank J. Lucks and M. Evans at Northwestern and R. Kretsch at Stanford for discussions. Some of this work was performed at the Stanford-SLAC Cryo-EM Center (S2C2), which is supported by the National Institutes of Health Common Fund Transformative High-Resolution Cryo-Electron Microscopy program (grant no. U24 GM129541). The US Government is authorized to reproduce and distribute reprints for Governmental purposes notwithstanding any copyright notation thereon. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the US Government or the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
D.S.K., M.C.J., A.W. and R.D. conceived the study and designed experiments. D.S.K., E.B., E.R. and C.K. worked on establishing the Evolink method. D.S.K. and E.B. carried out experiments for tethered ribosome evolution, orthogonal GFP expression and cellular growth rates. J.L. and K.J.S. carried out experiments for noncanonical amino acid incorporation. D.S.K. and A.W. analyzed results and designed the libraries. A.W. carried out computational modeling. V.T. performed chemical mapping experiments. D.S.K., V.T. and Y.L. performed cryo-EM sample preparation and data collection. V.T. and G.P. performed cryo-EM data analysis and structure determination. D.S.K., A.W., M.C.J. and R.D. wrote the manuscript with participation by all authors.
Corresponding author
Ethics declarations
Competing interests
M.C.J. and D.S.K. are coinventors on the US provisional patent application that incorporates discoveries described in this manuscript. M.C.J. has a financial interest in Pearl Bio, and his interests are reviewed and managed by Northwestern University in accordance with their competing interest policies. All other authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks Luc Jaeger, Jérôme Waldispühl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Optimization of molecular biology steps involved in library preparation workflow of Evolink.
a, A clonal sample of the tethered ribosome (Ribo-T v2) is linearized using different oligos compatible with multiple ligation protocols. b, From the different ligation products, generation of final amplicon for next-generation sequencing can happen with a wide range of ligation methods and starting template amounts in the PCR. Gel data representative of two independent experiments.
Extended Data Fig. 2 Enrichment of individual genotypes throughout full Evolink experiment.
a–c, Positively enriched genotypes (purple) and negative enriched genotypes (dark gray) can be tracked throughout multiple time points throughout selection. Genotypes that drop out during selection can also be identified (light gray). Corresponding heat maps that reveal trends in selected tether lengths also helped inform designs. Generally, across the three libraries tested in this work, (a) the Broad Sampling Library, (b) the Designed Junction Library, and (c) the Designed Junction + Length Refined Library, log2-fold enrichment values between -6 to 6 are observed. Enrichment and heatmap data representative of three independent experiments.
Extended Data Fig. 3 Score vs. Root-Mean-Standard-Deviation analysis of FARFAR2 simulations of enriched tether sequences.
a–d, For the Broad Sampling Library, we observe striking differences between simulations that constrained (blue) or did not constrain (orange) 3D structures of the Tether-H101 junction. Of the four modeled genotypes, two sequence (c,d) exhibit particularly substantial differences, hinting at structural instability in the Tether-H101 junction. e–h, When similar simulations are performed with enriched tether sequences from the Designed Junction Library (designed sequences at the Tether-H101 junction), the results of FARFAR2 simulations reach similarly low scores in constrained vs. unconstrained modeling runs.
Extended Data Fig. 4 Representative constrained and unconstrained 3D models of Designed Junction Library winner.
The winning genotype from Fig. 4h was modeled using Rosetta, and representative outputs are shown. In both the (a) unconstrained and (b) constrained model, the Designed Junction residues are predicted to base pair, reinforcing structural stability to this region.
Extended Data Fig. 5 Chemical reactivity of tethers to DMS in whole polysomes.
Targeted structure probing was performed on the tethers of both RiboTv2 and RiboTv3 polysomes via DMS-MaPseq. The per-nucleotide chemical reactivities of the tethers and their adjacent rRNA stems can be seen in the figure for both RiboTv2 (a) and RiboTv3 (b). Gray shaded nucleotides represent U and G residues that are not modified by DMS.
Extended Data Fig. 6 Raw Cryo-EM micrographs of Ribo-Tv3 polysomes.
Single-particle Cryo-EM was carried out on RiboTv3 polysomes. a, A representative raw micrograph shows that RiboTv3 polysomes look like characteristic ‘beads on a string’ as expected for actively translating ribosomes. b, A tethered ribosome that has dissociated from an mRNA looks like an open clamshell, as would be expected for a tethered ribosome. The large and small subunits are indicated by white arrows.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10 and Tables 1–7.
Source data
Source Data Fig. 2
Raw count data of genotypes from the Broad Sampling Library experiment.
Source Data Fig. 3
Raw count data of genotypes from Ribo-T junction library experiment.
Source Data Fig. 4
Raw count data of genotypes from Designed Junction Library experiment.
Source Data Fig. 5
Raw count data of genotypes from targeted Designed Junction Library experiment. Raw growth time series data. Raw GFP expression data. Raw mass spectrometry data.
Source Data Extended Data Fig. 1
Raw image of Evolink molecular biology validation gel.
Rights and permissions
About this article
Cite this article
Kim, D.S., Watkins, A., Bidstrup, E. et al. Three-dimensional structure-guided evolution of a ribosome with tethered subunits. Nat Chem Biol 18, 990–998 (2022). https://doi.org/10.1038/s41589-022-01064-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-022-01064-w
This article is cited by
-
Cell-free Biosynthesis of Peptidomimetics
Biotechnology and Bioprocess Engineering (2023)
-
Ribosome-mediated biosynthesis of pyridazinone oligomers in vitro
Nature Communications (2022)
-
Shackled ribosomes unleashed
Nature Chemical Biology (2022)