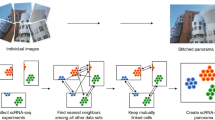
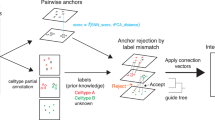
Abstract
Computational methods for integrating single-cell transcriptomic data from multiple samples and conditions do not generally account for imbalances in the cell types measured in different datasets. In this study, we examined how differences in the cell types present, the number of cells per cell type and the cell type proportions across samples affect downstream analyses after integration. The Iniquitate pipeline assesses the robustness of integration results after perturbing the degree of imbalance between datasets. Benchmarking of five state-of-the-art single-cell RNA sequencing integration techniques in 2,600 integration experiments indicates that sample imbalance has substantial impacts on downstream analyses and the biological interpretation of integration results. Imbalance perturbation led to statistically significant variation in unsupervised clustering, cell type classification, differential expression and marker gene annotation, query-to-reference mapping and trajectory inference. We quantified the impacts of imbalance through newly introduced properties—aggregate cell type support and minimum cell type center distance. To better characterize and mitigate impacts of imbalance, we introduce balanced clustering metrics and imbalanced integration guidelines for integration method users.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data necessary to reproduce the results of the study can be downloaded from Figshare: https://doi.org/10.6084/m9.figshare.24625302.v1.
The raw data for the datasets analyzed in this study can be found in the following links/accessions: two-batch balanced PBMC data8,9: SRP073767 and https://www.10xgenomics.com/resources/datasets/8-k-pbm-cs-from-a-healthy-donor-2-standard-2-1-0; two-batch and four-batch imbalanced PBMC data14: GSE132044; PDAC tumor data16—Genome Sequencing Archive: CRA001160; mouse hindbrain developmental data15: GSE118068; and mammalian organogenesis data10: GSE119945.
Code availability
All of the code necessary to reproduce the results of the Iniquitate pipeline is available at https://github.com/hsmaan/Iniquitate.
The vignette for a walkthrough of the imbalanced integration guidelines outlined in Results section “End-user guidelines for imbalanced integration” can be found in the Iniquitate documentation (https://github.com/hsmaan/Iniquitate/tree/main/docs). The Python package for implementing the balanced clustering metrics can be found at https://github.com/hsmaan/balanced-clustering.
References
Argelaguet, R. et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 576, 487–491 (2019).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Amezquita, R. A. et al. Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145 (2020).
Ming, J. et al. FIRM: flexible integration of single-cell RNA-sequencing data for large-scale multi-tissue cell atlas datasets. Brief. Bioinform. 23, bbac167 (2022).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
10x Genomics. 8k PBMCs from a healthy donor, single cell gene expression dataset by Cell Ranger 2.1.0. https://www.10xgenomics.com/resources/datasets/8-k-pbm-cs-from-a-healthy-donor-2-standard-2-1-0 (2017).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Abdelaal, T. et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 20, 194 (2019).
Clarke, Z. A. et al. Tutorial: guidelines for annotating single-cell transcriptomic maps using automated and manual methods. Nat. Protoc. 16, 2749–2764 (2021).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019).
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020).
Vladoiu, M. C. et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature 572, 67–73 (2019).
Peng, J. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 29, 725–738 (2019).
Polański, K. et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Hie, B., Bryson, B. & Berger, B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 37, 685–691 (2019).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Buitinck, L. et al. API design for machine learning software: experiences from the scikit-learn project. Preprint at https://doi.org/10.48550/arXiv.1309.0238 (2013).
Goutte, C. & Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In Advances in Information Retrieval 345–359. https://doi.org/10.1007/978-3-540-31865-1_25 (Springer, 2005).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Svensson, V., da Veiga Beltrame, E. & Pachter, L. A curated database reveals trends in single-cell transcriptomics. Database (Oxford) 2020, baaa073 (2020).
Dohmen, J. et al. Identifying tumor cells at the single-cell level using machine learning. Genome Biol. 23, 123 (2022).
Trinh, M. K. et al. Precise identification of cancer cells from allelic imbalances in single cell transcriptomes. Commun. Biol. 5, 884 (2022).
Xu, Y., Liu, J., Nipper, M. & Wang, P. Ductal vs. acinar? Recent insights into identifying cell lineage of pancreatic ductal adenocarcinoma. Ann. Pancreat. Cancer 2, 11 (2019).
Backx, E. et al. On the origin of pancreatic cancer: molecular tumor subtypes in perspective of exocrine cell plasticity. Cell Mol. Gastroenterol. Hepatol. 13, 1243–1253 (2022).
Hubert, L. & Arabie, P. Comparing partitions. J. Classif. 2, 193–218 (1985).
Vinh, N. X., Epps, J. & Bailey, J. Information theoretic measures for clusterings comparison: variants, properties, normalization and correction for chance. J. Mach. Learn. Res. 11, 2837–2854 (2010).
Argelaguet, R., Cuomo, A. S., Stegle, O. & Marioni, J. C. Computational principles and challenges in single-cell data integration. Nat. Biotechnol. 39, 1202–1215 (2021).
Ogbeide, S., Giannese, F., Mincarelli, L. & Macaulay, I. C. Into the multiverse: advances in single-cell multiomic profiling. Trends Genet. 38, 831–843 (2022).
Andreatta, M. & Carmona, S. J. STACAS: sub-type anchor correction for alignment in Seurat to integrate single-cell RNA-seq data. Bioinformatics 37, 882–884 (2021).
Johansen, N. & Quon, G. ScAlign: a tool for alignment, integration, and rare cell identification from scRNA-seq data. Genome Biol. 20, 166 (2019).
Hu, Z., Ahmed, A. A. & Yau, C. CIDER: an interpretable meta-clustering framework for single-cell RNA-seq data integration and evaluation. Genome Biol. 22, 337 (2021).
Demetçi, P., Santorella, R., Sandstede, B. & Singh, R. Unsupervised integration of single-cell multi-omics datasets with disproportionate cell-type representation. Preprint at bioRxiv https://doi.org/10.1101/2021.11.09.467903 (2022).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337 (2019).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Chijimatsu, R. et al. Establishment of a reference single-cell RNA sequencing dataset for human pancreatic adenocarcinoma. iScience 25, 104659 (2022).
Tickle, T., Tirosh, I., Georgescu, C., Brown, M. & Haas, B. Infer copy number variation from single-cell RNA-seq data. https://doi.org/doi:10.18129/B9.bioc.infercnv (2019).
Steele, N. G. et al. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat. Cancer 1, 1097–1112 (2020).
Chen, K. et al. Immune profiling and prognostic model of pancreatic cancer using quantitative pathology and single-cell RNA sequencing. J. Transl. Med. 21, 210 (2023).
Welch, J. D. et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887 (2019).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://doi.org/10.48550/arXiv.1802.03426 (2018).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019).
Haghverdi, L., Büttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Winer, B. J., Brown, D. R. & Michels, K. M. Statistical Principles in Experimental Design 3rd edn (McGraw-Hill, 1991).
Rosenberg, A. & Hirschberg, J. V-Measure: a conditional entropy-based external cluster evaluation measure. Proc. of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning 410–420 (Association for Computational Linguistics, 2007).
Acknowledgements
We would like to thank members of the Bo Wang and Kieran Campbell laboratories for insightful discussion and feedback on this work. H.M. is supported by a doctoral fellowship from the Natural Sciences and Engineering Research Council of Canada (NSERC). M.G. is supported by the Princess Margaret Cancer Foundation and a Health Informatics and Data Science award from the Terry Fox Research Institute. C.Y. and M.G are supported by University of Toronto Data Science Institute doctoral fellowships. This work was supported by funding from the Canadian Institutes of Health Research (project grant PJT175270, to K.C.), funding from the NSERC (RGPIN-2020-04083, to K.C., and RGPIN-2020-06189 and DGECR-2020-00294, to B.W.), a Canada Foundation for Innovation John R. Evans Leaders Fund award (to K.C.), the CIFAR AI Chairs Program (to B.W.) and the Peter Munk Cardiac Centre AI Fund at the University Health Network (to B.W.). This research was undertaken, in part, thanks to funding from the Canada Research Chairs Program. Figures 1, 4a and 6 were created with BioRender.
Author information
Authors and Affiliations
Contributions
H.M., K.C. and B.W. conceptualized the ideas and experiments. H.M. performed the manuscript experiments and associated analysis. H.M. and L.Z. performed the statistical analyses accompanying the experiments. C.Y. and M.G. processed and annotated the PDAC data and helped with associated experiments and analysis. H.M. developed the balanced metrics and performed associated analysis. H.M. developed the guidelines for imbalanced integration. K.C. and B.W. funded the project and provided supervision. H.M. wrote the original manuscript, and all authors provided review and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
B.W. is on the Strategic Advisory Board of Vevo Therapeutics, Inc. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–7 and Supplementary Figs. 1–36.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Maan, H., Zhang, L., Yu, C. et al. Characterizing the impacts of dataset imbalance on single-cell data integration. Nat Biotechnol (2024). https://doi.org/10.1038/s41587-023-02097-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-023-02097-9