Abstract
Cellular sodium ion (Na+) homeostasis is integral to organism physiology. Our current understanding of Na+ homeostasis is largely limited to Na+ transport at the plasma membrane. Organelles may also contribute to Na+ homeostasis; however, the direction of Na+ flow across organelle membranes is unknown because organellar Na+ cannot be imaged. Here we report a pH-independent, organelle-targetable, ratiometric probe that reports lumenal Na+. It is a DNA nanodevice containing a Na+-sensitive fluorophore, a reference dye and an organelle-targeting domain. By measuring Na+ at single endosome resolution in mammalian cells and Caenorhabditis elegans, we discovered that lumenal Na+ levels in each stage of the endolysosomal pathway exceed cytosolic levels and decrease as endosomes mature. Further, we find that lysosomal Na+ levels in nematodes are modulated by the Na+/H+ exchanger NHX-5 in response to salt stress. The ability to image subcellular Na+ will unveil mechanisms of Na+ homeostasis at an increased level of cellular detail.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The raw data supporting Figs. 1–4 are available for public access at Figshare: https://doi.org/10.6084/m9.figshare.23938503 (ref. 67). Source data are provided with this paper.
References
Shapovalov, G. et al. Organelle membrane derived patches: reshaping classical methods for new targets. Sci. Rep. 7, 14082 (2017).
Nass, R., Cunningham, K. W. & Rao, R. Intracellular sequestration of sodium by a novel Na+/H+ exchanger in yeast is enhanced by mutations in the plasma membrane H+-ATPase. Insights into mechanisms of sodium tolerance. J. Biol. Chem. 272, 26145–26152 (1997).
Nass, R. & Rao, R. Novel localization of a Na+/H+ exchanger in a late endosomal compartment of yeast. Implications for vacuole biogenesis. J. Biol. Chem. 273, 21054–21060 (1998).
Orlowski, J. & Grinstein, S. Na+/H+ exchangers. Compr. Physiol. 1, 2083–2100 (2011).
Pedersen, S. F. & Counillon, L. The SLC9A-C mammalian Na+/H+ exchanger family: molecules, mechanisms, and physiology. Physiol. Rev. 99, 2015–2113 (2019).
Kondapalli, K. C. et al. Functional evaluation of autism-associated mutations in NHE9. Nat. Commun. 4, 2510 (2013).
Morrow, E. M. et al. Identifying autism loci and genes by tracing recent shared ancestry. Science 321, 218–223 (2008).
Pescosolido, M. F., Ouyang, Q., Liu, J. S. & Morrow, E. M. Loss of Christianson syndrome Na+/H+ exchanger 6 (NHE6) causes abnormal endosome maturation and trafficking underlying lysosome dysfunction in neurons. J. Neurosci. 41, 9235–9256 (2021).
Fukada-Tanaka, S., Inagaki, Y., Yamaguchi, T., Saito, N. & Iida, S. Colour-enhancing protein in blue petals. Nature 407, 581 (2000).
Lamason, R. L. et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786 (2005).
Cang, C., Bekele, B. & Ren, D. The voltage-gated sodium channel TPC1 confers endolysosomal excitability. Nat. Chem. Biol. 10, 463–469 (2014).
Cang, C. et al. mTOR regulates lysosomal ATP-sensitive two-pore Na+ channels to adapt to metabolic state. Cell 152, 778–790 (2013).
Wang, X. et al. TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383 (2012).
Saminathan, A. et al. A DNA-based voltmeter for organelles. Nat. Nanotechnol. 16, 96–103 (2021).
Erecińska, M. & Silver, I. A. Ions and energy in mammalian brain. Prog. Neurobiol. 43, 37–71 (1994).
Harootunian, A. T., Kao, J. P., Eckert, B. K. & Tsien, R. Y. Fluorescence ratio imaging of cytosolic free Na+ in individual fibroblasts and lymphocytes. J. Biol. Chem. 264, 19458–19467 (1989).
Minta, A. & Tsien, R. Y. Fluorescent indicators for cytosolic sodium. J. Biol. Chem. 264, 19449–19457 (1989).
Steinberg, B. E. et al. A cation counterflux supports lysosomal acidification. J. Cell Biol. 189, 1171–1186 (2010).
Leung, K., Chakraborty, K., Saminathan, A. & Krishnan, Y. A DNA nanomachine chemically resolves lysosomes in live cells. Nat. Nanotechnol. 14, 176–183 (2019).
Zhu, H. et al. Metabolomic profiling of single enlarged lysosomes. Nat. Methods 18, 788–798 (2021).
Martin, V. V., Rothe, A. & Gee, K. R. Fluorescent metal ion indicators based on benzoannelated crown systems: a green fluorescent indicator for intracellular sodium ions. Bioorg. Med. Chem. Lett. 15, 1851–1855 (2005).
Holmehave, J., Pedersen, S. K., Jensen, H. & Ogilby, P. R. Aarhus green: a tetrafluoro-substituted derivative of fluorescein. Arkivoc 2015, 52 (2015).
Rostovtsev, V. V., Green, L. G., Fokin, V. V. & Sharpless, K. B. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ‘ligation’ of azides and terminal alkynes. Angew. Chem. Int. Ed. 41, 2596–2599 (2002).
Chang, P. V. et al. Copper-free click chemistry in living animals. Proc. Natl Acad. Sci. USA 107, 1821–1826 (2010).
Veetil, A. T. et al. DNA-based fluorescent probes of NOS2 activity in live brains. Proc. Natl Acad. Sci. USA 117, 14694–14702 (2020).
Krishnan, Y., Zou, J. & Jani, M. S. Quantitative imaging of biochemistry in situ and at the nanoscale. ACS Cent. Sci. 6, 1938–1954 (2020).
Surana, S., Bhat, J. M., Koushika, S. P. & Krishnan, Y. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat. Commun. 2, 340 (2011).
Suresh, B. et al. Tubular lysosomes harbor active ion gradients and poise macrophages for phagocytosis. Proc. Natl Acad. Sci. USA 118, e2113174118 (2021).
Cui, C. et al. A lysosome-targeted DNA nanodevice selectively targets macrophages to attenuate tumours. Nat. Nanotechnol. 16, 1394–1402 (2021).
Dan, K., Veetil, A. T., Chakraborty, K. & Krishnan, Y. DNA nanodevices map enzymatic activity in organelles. Nat. Nanotechnol. 14, 252–259 (2019).
Surana, S., Bhatia, D. & Krishnan, Y. A method to study in vivo stability of DNA nanostructures. Methods 64, 94–100 (2013).
Ishiguro, H., Steward, M. C., Lindsay, A. R. & Case, R. M. Accumulation of intracellular HCO3- by Na+-HCO3- cotransport in interlobular ducts from guinea-pig pancreas. J. Physiol. (Lond.) 495, 169–178 (1996).
Saminathan, A., Zajac, M., Anees, P. & Krishnan, Y. Organelle-level precision with next-generation targeting technologies. Nat. Rev. Mater. 7, 355–371 (2022).
Saha, S., Prakash, V., Halder, S., Chakraborty, K. & Krishnan, Y. A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 10, 645–651 (2015).
Narayanaswamy, N. et al. A pH-correctable, DNA-based fluorescent reporter for organellar calcium. Nat. Methods 16, 95–102 (2019).
Calcraft, P. J. et al. NAADP mobilizes calcium from acidic organelles through two-pore channels. Nature 459, 596–600 (2009).
Ogunbayo, O. A. et al. mTORC1 controls lysosomal Ca2+ release through the two-pore channel TPC2. Sci. Signal. 11, eaao5775 (2018).
Kang, Y.-L. et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. Proc. Natl Acad. Sci. USA 117, 20803–20813 (2020).
Prasad, H. & Rao, R. The Na+/H+ exchanger NHE6 modulates endosomal pH to control processing of amyloid precursor protein in a cell culture model of Alzheimer disease. J. Biol. Chem. 290, 5311–5327 (2015).
Yang, Y. & Guo, Y. Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804 (2018).
Lamitina, S. T., Morrison, R., Moeckel, G. W. & Strange, K. Adaptation of the nematode Caenorhabditis elegans to extreme osmotic stress. Am. J. Physiol. Cell Physiol. 286, C785–C791 (2004).
Urso, S. J. & Lamitina, T. The C. elegans hypertonic stress response: big insights from shrinking worms. Cell. Physiol. Biochem. 55, 89–105 (2021).
Nehrke, K. & Melvin, J. E. The NHX family of Na+-H+ exchangers in Caenorhabditis elegans. J. Biol. Chem. 277, 29036–29044 (2002).
Kim, J. et al. NHX-5, an endosomal Na+/H+ exchanger, is associated with metformin action. J. Biol. Chem. 291, 18591–18599 (2016).
Collins, J. F. et al. Molecular cloning, sequencing, tissue distribution, and functional expression of a Na+/H+ exchanger (NHE-2). Proc. Natl Acad. Sci. USA 90, 3938–3942 (1993).
Oberheide, K., Puchkov, D. & Jentsch, T. J. Loss of the Na+/H+ exchanger NHE8 causes male infertility in mice by disrupting acrosome formation. J. Biol. Chem. 292, 10845–10854 (2017).
Assaha, D. V. M., Ueda, A., Saneoka, H., Al-Yahyai, R. & Yaish, M. W. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 8, 509 (2017).
Mager, T., Rimon, A., Padan, E. & Fendler, K. Transport mechanism and pH regulation of the Na+/H+ antiporter NhaA from Escherichia coli: an electrophysiological study. J. Biol. Chem. 286, 23570–23581 (2011).
Mindell, J. A. Lysosomal acidification mechanisms. Annu. Rev. Physiol. 74, 69–86 (2012).
de Voer, G., Peters, D. & Taschner, P. E. M. Caenorhabditis elegans as a model for lysosomal storage disorders. Biochim. Biophys. Acta 1782, 433–446 (2008).
Chakraborty, K., Leung, K. & Krishnan, Y. High lumenal chloride in the lysosome is critical for lysosome function. eLife 6, e28862 (2017).
Rebsamen, M. et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519, 477–481 (2015).
Ba, Q., Raghavan, G., Kiselyov, K. & Yang, G. Whole-cell scale dynamic organization of lysosomes revealed by spatial statistical analysis. Cell Rep. 23, 3591–3606 (2018).
Platt, C. D. et al. Leucine-rich repeat containing 8A (LRRC8A)-dependent volume-regulated anion channel activity is dispensable for T-cell development and function. J. Allergy Clin. Immunol. 140, 1651–1659 (2017).
López-Hernández, T., Puchkov, D., Krause, E., Maritzen, T. & Haucke, V. Endocytic regulation of cellular ion homeostasis controls lysosome biogenesis. Nat. Cell Biol. 22, 815–827 (2020).
Jeffery, J. & Jörnvall, H. Enzyme relationships in a sorbitol pathway that bypasses glycolysis and pentose phosphates in glucose metabolism. Proc. Natl Acad. Sci. USA 80, 901–905 (1983).
Burg, M. B., Ferraris, J. D. & Dmitrieva, N. I. Cellular response to hyperosmotic stresses. Physiol. Rev. 87, 1441–1474 (2007).
Galluzzi, L., Pietrocola, F., Levine, B. & Kroemer, G. Metabolic control of autophagy. Cell 159, 1263–1276 (2014).
Kandasamy, P., Gyimesi, G., Kanai, Y. & Hediger, M. A. Amino acid transporters revisited: new views in health and disease. Trends Biochem. Sci. 43, 752–789 (2018).
Wang, S. et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347, 188–194 (2015).
Efeyan, A., Comb, W. C. & Sabatini, D. M. Nutrient-sensing mechanisms and pathways. Nature 517, 302–310 (2015).
Lamming, D. W. & Bar-Peled, L. Lysosome: the metabolic signaling hub. Traffic 20, 27–38 (2019).
Lord, S. J., Velle, K. B., Mullins, R. D. & Fritz-Laylin, L. K. SuperPlots: communicating reproducibility and variability in cell biology. J. Cell Biol. 219, e202001064 (2020).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Strømme, P. et al. X-linked Angelman-like syndrome caused by Slc9a6 knockout in mice exhibits evidence of endosomal-lysosomal dysfunction. Brain J. Neurol. 134, 3369–3383 (2011).
Nasser, H. et al. Establishment of bone marrow-derived M-CSF receptor-dependent self-renewing macrophages. Cell Death Discov. 6, 1–14 (2020).
Zou, J. et al. DNA nanodevice for mapping sodium at single organelle resolution. Datasets. Figshare. https://doi.org/10.6084/m9.figshare.23938503 (2023).
Acknowledgements
We thank E. Perozo (University of Chicago), G. Ruvkun (Massachusetts General Hospital), A. Concepcion (University of Chicago) and A. L. Chun for valuable discussions and input on the manuscript. We thank C. Labno at the integrated light microscopy facilities at the University of Chicago for technical help and T. Wu for assistance with qRT–PCR. We thank K. Nehrke (University of Rochester Medical Center) for sharing NHX-5::GFP plasmid. Y.K. acknowledges funding from NIH grants 1DP1GM149751-01, 1R01NS112139-01A1, 1R21NS114428-01, 1R21HL161825-01A1 and 1R01GM147197-01 (to Y.K. and R.R.); FA9550-19-0003 from the AFOSR; HFSP grant RGP0032/2022 and the Ono Pharma Foundation.
Author information
Authors and Affiliations
Contributions
J.Z. and Y.K. designed every aspect of the study. K.M., A.T.V. and J.R. designed and synthesized the CG dye. J.Z. made, characterized and validated RatiNa in vitro, in cells and in vivo. J.Z. and P.A. performed Na+ measurements in worms. D.O. performed all the brood size experiments. P.A. performed pH measurements in worms. J.J.S. and P.K. made the NHX-5::GFP worm. P.D.G. and R.R. provided NHE6 KO macrophages. J.Z. and Y.K. analyzed and interpreted all data. J.Z., K.M. and Y.K. wrote the paper. All authors provided input on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. Y.K. is a cofounder of Esya Inc and MacroLogic Inc, which use DNA nanodevices to develop diagnostics and therapeutics, respectively.
Peer review
Peer review information
Nature Biotechnology thanks Haoxing Xu for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
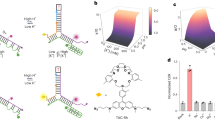
Extended Data Fig. 1 Chicago green (CG) is a pH insensitive and Na+ selective fluorophore.
a, Na+ sensing mechanism of CG b, Excitation (black) and emission (green) spectra of 100 nM CG increases with increasing Na+. c, Dissociation constant (Kd) of CG for Na+ does not vary with pH from pH 4.5–7.4. d, Individual in vitro calibration profiles of RatiNa at different pH in Fig. 1d. e, Kd of RatiNa for Na+ at different pH values as calculated from d. Kd of RatiNa is invariant from pH 4.5–7.4. f, RatiNa Na+ calibration profile from 1 mM to 200 mM Na+ in linear scale. g, RatiNa yields a Kd of 4.5 M for K+ and is 27-times more selective for Na+. Data in c-g are presented as mean values ± s.d. from n = 3 independent experiments.
Extended Data Fig. 2 Calibration of RatiNa and its stability in lysosomes of RAW 264.7 macrophages.
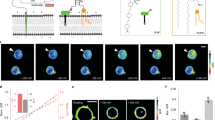
a, Lysosomes in RAW 264.7 macrophages prelabeled with TMR-dextran (cyan) and imaged at different chase times of RatiNaAT (magenta). Images are representative from n = 2 independent experiments. b, Histogram of intensity ratios of RatiNaAT/TMR-dextran (R/G) in single lysosomes at different chase times. Decreasing R/G indicates more DNA degradation. c, DNA degradation as a function of time quantified from b. Data are presented as mean values ± s.e.m. from n = 2 independent experiments. d, Normalized RatiNa signal (G/R) of n = 127 single lysosome in RAW macrophages versus the normalizing dye signal (R). No correlation between G/R and R, indicates RatiNa output depends only on Na+ level and independent of probe concentration. e, Fluorescence images of RatiNaAT labeled RAW 264.7 macrophages in CG (G) and ATTO (R) channels. Representative images from n = 3 independent experiments. f, Images of RatiNa-labeled lysosomes clamped at high and low Na+ of 145 mM and 5 mM in native lysosomes. G/R heat maps show RatiNa response in native lysosomes. Representative images from n = 3 independent experiments. g, Histogram of G/R values of RatiNa-labeled single lysosomes. Despite autofluorescence in each system, the fold change in G/R signal of RatiNa in lysosomes of RAW 264.7 macrophages is comparable to that in C. elegans and on beads. h, Schematic of workflow from raw images to Na+ heat maps of single organelles. Fluorescent images in the CG (G) and ATTO (R) channels are used to construct the G/R image. Then the Na+ heatmap was generated from the calibration curve of G/R to [Na+].
Extended Data Fig. 3 RatiNa is targeted specifically to each organelle on the endolysosomal pathway.
a, Representative images of C. elegans coelomocytes reveal negligible off-target labeling between RatiNaAT and indicated endocytic markers and chase times. b, RatiNa is targeted to a specific endocytic organelle at fixed chase time. Colocalization is calculated as percentage of organelles having both lumenal RatiNaAT and membrane marker fluorescence over all RatiNaAT containing organelles. Data are presented as mean values ± s.d. from n = 3 independent experiments. (n = 9 coelomocytes, 6 worms for 5 min chase of RAB-5::GFP; n = 7 coelomocytes, 6 worms for 17 min chase of RAB-5::GFP; n = 4 coelomocytes, 4 worms for 5 min chase of RAB-7::GFP; n = 6 coelomocytes, 5 worms for 17 min chase of RAB-7::GFP; n = 7 coelomocytes, 6 worms for 17 min chase of LMP-1::GFP; n = 9 coelomocytes, 6 worms for 60 min chase of LMP-1::GFP).
Extended Data Fig. 4 Na+ transporter mutants are less resistant to high salt stress and show elevated lysosomal Na+.
a, Brood sizes of Na+ transporter mutants upon high salt stress. Na+ transporter deletion mutants cannot adapt to 400 mM NaCl unlike WT worms. Arrowheads indicate points of lysosomal Na+ measurement that is, worms acutely stressed (Ac) with 200 mM. Data are presented from n = 3 independent experiments. b, Representative Na+ heatmaps of Na+ transporter deletion mutant worms. c, qRT-PCR shows that mRNA expression level of Na+ transporters do not change appreciably upon salt stress in N2 worms. Fold change in mRNA levels of Na+ transporters between Ac and Ch conditions is shown. act-1 was used as reference gene. Data are from n = 2–3 independent experiments. d. Lysosomal Na+ levels of Na+ transporter mutant worms under unstressed (Ns) and acutely stressed (Ac) condition. High lysosomal Na+ is observed in all investigated Na+ transporter mutants: 9 mM in Ns and 63 mM in Ac for nhx-5(−) worms. 24 mM in Ns and 51 mM in Ac for nhx-7(−) worms. 34 mM in Ns and 55 mM in Ac for nhx-8(−) worms. 25 mM in Ns and 36 mM in Ac for ncx-2(−) worms. Two sample two-tailed t test was used for statistical analysis assuming equal variance. ***P = 2.5 × 10−13, ***P = 1.7 × 10−10, ***P = 2.2 × 10−5, *P = 0.016 for nhx-5(−), nhx-7(−), nhx-8(−), ncx-2(−). All error bars are presented as mean values ± s.d.
Extended Data Fig. 5 Lysosomal pH of salt stressed worms.
a. PAGE analysis of the I-switch-based pH reporter module denoted Br-I-switch19. DD strand has Alexa488 as a donor, DA strand has Alexa647 as acceptor. b. pH calibration curve of Br-I-switch shows ~20 fold change of D/A signal from pH 5.0 to 6.0, with highest sensitivity near pH 5.5, the pH of coelomocyte lysosomes35. Data are presented as mean values ± s.d. from n = 3 independent experiments c. pH measurement of single lysosome of N2 and nhx-5(−) worms in unstressed (Ns) and acutely salt stressed (Ac) conditions. Ns and Ac nhx-5(−) worms show lower pH. Error bar represents ± s.d. Two sample two-tailed t-test was used for statistical analysis assuming equal variance. ***P = 1.4 × 10-120, ***P = 7.4 × 10-88, P = 0.25, *P = 0.042 for N2 Ns to N2 Ac, nhx-5(−) Ns to nhx-5(−) Ac, N2 Ns to nhx-5(−) Ns, N2 Ac to nhx-5(−) Ac. d. Representative images of Br-I-switch in Donor (D), Acceptor (A) FRET (D/A) channels and respective pH heatmaps.
Extended Data Fig. 6 Mutants lacking key lysosomal genes adapt to high salt stress differently.
a, Brood sizes of deletion mutants of lysosomal storage disorder genes. Data are presented as mean values from n = 3 independent experiments. gba-3(−) and ctns-1(−), clh-6(−) and XT7 (cln-3.2(−);cln3.3(−); cln-3.1(−)) are worm models Gaucher’s disease, Cystinosis, osteopetrosis and Batten’s disease respectively, and more susceptible to chronic stress than N2 worms. b, A deletion mutant of F13H10.3, the C. elegans homolog of human SLC38A9, denoted (slc38a9(−)) is more susceptible to Ac salt stress. Brood sizes are presented as mean values from n = 3 independent experiments. Arrows indicate points of lysosomal Na+ measurement that is, worms acutely stressed (Ac) with 200 mM. c, Representative Na+ heatmaps of slc38a9(−) worms in Ns and Ac. d, Lysosomal Na+ measurements in the indicated slc38a9(−) worms with and without salt stress. Higher lysosomal Na+ level of 52 mM in Ac worms compared to 39 mM in Ns worms, Error bar represents ± s.d. Two sample two-tailed t-test was used for statistical analysis assuming equal variance. P = 0.4, ***P = 6.8 × 10−4 for N2 Ns to slc38a9(−) Ns, slc38a9(−) Ns to slc38a9(−) Ac.
Supplementary information
Supplementary Information
Supplementary Figs. 1–10, Supplementary Notes 1–12 and Supplementary Table 1.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 5
Unprocessed PAGE gel.
Source Data Extended Data Fig. 6
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zou, J., Mitra, K., Anees, P. et al. A DNA nanodevice for mapping sodium at single-organelle resolution. Nat Biotechnol (2023). https://doi.org/10.1038/s41587-023-01950-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-023-01950-1
This article is cited by
-
Organellar electrophysiology: DNA nanodevices charging at the unmeasured
BMC Biology (2024)
-
DNA nanodevices map intracellular ions
Nature Biotechnology (2023)