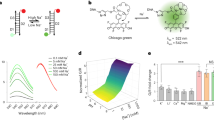
Abstract
Cell surface potassium ion (K+) channels regulate nutrient transport, cell migration and intercellular communication by controlling K+ permeability and are thought to be active only at the plasma membrane. Although these channels transit the trans-Golgi network, early and recycling endosomes, whether they are active in these organelles is unknown. Here we describe a pH-correctable, ratiometric reporter for K+ called pHlicKer, use it to probe the compartment-specific activity of a prototypical voltage-gated K+ channel, Kv11.1, and show that this cell surface channel is active in organelles. Lumenal K+ in organelles increased in cells expressing wild-type Kv11.1 channels but not after treatment with current blockers. Mutant Kv11.1 channels, with impaired transport function, failed to increase K+ levels in recycling endosomes, an effect rescued by pharmacological correction. By providing a way to map the organelle-specific activity of K+ channels, pHlicKer technology could help identify new organellar K+ channels or channel modulators with nuanced functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All data related to the study are included in the article and supporting information. The raw data supporting Figs. 1–6, extended data figures and supplementary figures, respectively, are available for public access at figshare54,55,56,57,58,59,60,61(https://figshare.com/articles/dataset/Figure_1/23713359, https://figshare.com/articles/dataset/Figure_3/23713776, https://figshare.com/articles/dataset/Figure_4/23713821, https://figshare.com/articles/dataset/Figure_5/23713833, https://figshare.com/articles/dataset/Figure_6/23713851, https://figshare.com/articles/dataset/Extended_data_figures/23713899 and https://figshare.com/articles/dataset/Supplementary_Figures/23713947). Source data are provided with this paper.
References
Foo, B., Williamson, B., Young, J. C., Lukacs, G. & Shrier, A. hERG quality control and the long QT syndrome. J. Physiol. 594, 2469–2481 (2016).
Saminathan, A. et al. A DNA-based voltmeter for organelles. Nat. Nanotechnol. 16, 96–103 (2021).
Irannejad, R. et al. Conformational biosensors reveal GPCR signalling from endosomes. Nature 495, 534–538 (2013).
Tao, X., Avalos, J. L., Chen, J. & MacKinnon, R. Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution. Science 326, 1668–1674 (2009).
Sambath, K. et al. Potassium ion fluorescence probes: structures, properties and bioimaging. ChemPhotoChem 5, 317–325 (2021).
He, H., Mortellaro, M. A., Leiner, M. J. P., Fraatz, R. J. & Tusa, J. K. A fluorescent sensor with high selectivity and sensitivity for potassium in water. J. Am. Chem. Soc. 125, 1468–1469 (2003).
Padmawar, P., Yao, X., Bloch, O., Manley, G. T. & Verkman, A. S. K+ waves in brain cortex visualized using a long-wavelength K+-sensing fluorescent indicator. Nat. Methods 2, 825–827 (2005).
Schapiro, F. B. & Grinstein, S. Determinants of the pH of the Golgi complex. J. Biol. Chem. 275, 21025–21032 (2000).
Reeves, E. P. et al. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416, 291–297 (2002).
Chakraborty, K., Veetil, A. T., Jaffrey, S. R. & Krishnan, Y. Nucleic acid-based nanodevices in biological imaging. Annu. Rev. Biochem. 85, 349–373 (2016).
Saminathan, A., Zajac, M., Anees, P. & Krishnan, Y. Organelle-level precision with next-generation targeting technologies. Nat. Rev. Mater. 7, 355–371 (2022).
Krishnan, Y., Zou, J. & Jani, M. S. Quantitative imaging of biochemistry in situ and at the nanoscale. ACS Cent. Sci. 6, 1938–1954 (2020).
Modi, S. et al. A DNA nanomachine that maps spatial and temporal pH changes inside living cells. Nat. Nanotechnol. 4, 325–330 (2009).
Modi, S., Nizak, C., Surana, S., Halder, S. & Krishnan, Y. Two DNA nanomachines map pH changes along intersecting endocytic pathways inside the same cell. Nat. Nanotechnol. 8, 459–467 (2013).
Narayanaswamy, N. et al. A pH-correctable, DNA-based fluorescent reporter for organellar calcium. Nat. Methods 16, 95–102 (2019).
Saha, S., Prakash, V., Halder, S., Chakraborty, K. & Krishnan, Y. A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells. Nat. Nanotechnol. 10, 645–651 (2015).
Zhou, Z. et al. Properties of HERG channels stably expressed in HEK 293 cells studied at physiological temperature. Biophys. J. 74, 230–241 (1998).
Surana, S., Shenoy, A. R. & Krishnan, Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol. 10, 741–747 (2015).
Surana, S., Bhat, J. M., Koushika, S. P. & Krishnan, Y. An autonomous DNA nanomachine maps spatiotemporal pH changes in a multicellular living organism. Nat. Commun. 2, 340 (2011).
Bhatia, D., Surana, S., Chakraborty, S., Koushika, S. P. & Krishnan, Y. A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging. Nat. Commun. 2, 339 (2011).
Veetil, A. T. et al. Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat. Nanotechnol. 12, 1183–1189 (2017).
Leung, K., Chakraborty, K., Saminathan, A. & Krishnan, Y. A DNA nanomachine chemically resolves lysosomes in live cells. Nat. Nanotechnol. 14, 176–183 (2019).
Jani, M. S., Zou, J., Veetil, A. T. & Krishnan, Y. A DNA-based fluorescent probe maps NOS3 activity with subcellular spatial resolution. Nat. Chem. Biol. 16, 660–666 (2020).
Osei-Owusu, J. et al. Proton-activated chloride channel PAC regulates endosomal acidification and transferrin receptor-mediated endocytosis. Cell Rep. 34, 108683 (2021).
Di, A. et al. The TWIK2 potassium efflux channel in macrophages mediates NLRP3 inflammasome-induced inflammation. Immunity 49, 56–65.e4 (2018).
Huang, L. S. et al. Endosomal trafficking of two pore K+ efflux channel TWIK2 to plasmalemma mediates NLRP3 inflammasome activation and inflammatory injury. eLife 12, e83842 (2023).
Sanguinetti, M. C., Jiang, C., Curran, M. E. & Keating, M. T. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 81, 299–307 (1995).
Keating, M. T. & Sanguinetti, M. C. Molecular and cellular mechanisms of cardiac arrhythmias. Cell 104, 569–580 (2001).
Sanguinetti, M. C. & Tristani-Firouzi, M. hERG potassium channels and cardiac arrhythmia. Nature 440, 463–469 (2006).
Zhou, Z., Gong, Q. & January, C. T. Correction of defective protein trafficking of a mutant HERG potassium channel in human long QT syndrome. Pharmacological and temperature effects. J. Biol. Chem. 274, 31123–31126 (1999).
Anderson, C. L. et al. Large-scale mutational analysis of Kv11.1 reveals molecular insights into type 2 long QT syndrome. Nat. Commun. 5, 5535 (2014).
Maxfield, F. R. & McGraw, T. E. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5, 121–132 (2004).
Hardel, N., Harmel, N., Zolles, G., Fakler, B. & Klöcker, N. Recycling endosomes supply cardiac pacemaker channels for regulated surface expression. Cardiovasc. Res. 79, 52–60 (2008).
Mohammad, S., Zhou, Z., Gong, Q. & January, C. T. Blockage of the HERG human cardiac K+ channel by the gastrointestinal prokinetic agent cisapride. Am. J. Physiol. 273, H2534–H2538 (1997).
Nakamura, N., Tanaka, S., Teko, Y., Mitsui, K. & Kanazawa, H. Four Na+/H+ exchanger isoforms are distributed to Golgi and post-Golgi compartments and are involved in organelle pH regulation. J. Biol. Chem. 280, 1561–1572 (2005).
Kellokumpu, S. Golgi ph, ion and redox homeostasis: how much do they really matter? Front. Cell Dev. Biol. 7, 93 (2019).
Solé, L. & Tamkun, M. M. Trafficking mechanisms underlying Nav channel subcellular localization in neurons. Channels 14, 1–17 (2020).
Smith, J. L. et al. Pharmacological correction of long QT-linked mutations in KCNH2 (hERG) increases the trafficking of Kv11.1 channels stored in the transitional endoplasmic reticulum. Am. J. Physiol. Cell Physiol. 305, C919–C930 (2013).
Ficker, E., Obejero-Paz, C. A., Zhao, S. & Brown, A. M. The binding site for channel blockers that rescue misprocessed human long QT syndrome type 2 ether-a-gogo-related gene (HERG) mutations. J. Biol. Chem. 277, 4989–4998 (2002).
Qile, M. et al. LUF7244 plus dofetilide rescues aberrant Kv11.1 trafficking and produces functional IKv11.1. Mol. Pharmacol. 97, 355–364 (2020).
Bischof, H. et al. Novel genetically encoded fluorescent probes enable real-time detection of potassium in vitro and in vivo. Nat. Commun. 8, 1422 (2017).
Bezanilla, F. The voltage sensor in voltage-dependent ion channels. Physiol. Rev. 80, 555–592 (2000).
Bian, J., Cui, J. & McDonald, T. V. HERG K+ channel activity is regulated by changes in phosphatidyl inositol 4,5-bisphosphate. Circ. Res. 89, 1168–1176 (2001).
Rodriguez, N. et al. Phosphatidylinositol-4,5-bisphosphate (PIP(2)) stabilizes the open pore conformation of the Kv11.1 (hERG) channel. Biophys. J. 99, 1110–1118 (2010).
Tan, X., Thapa, N., Choi, S. & Anderson, R. A. Emerging roles of PtdIns(4,5)P2–beyond the plasma membrane. J. Cell Sci. 128, 4047–4056 (2015).
Zhang, X., Hu, M., Yang, Y. & Xu, H. Organellar TRP channels. Nat. Struct. Mol. Biol. 25, 1009–1018 (2018).
Xu, H., Martinoia, E. & Szabo, I. Organellar channels and transporters. Cell Calcium 58, 1–10 (2015).
Prakash, V. et al. Quantitative mapping of endosomal DNA processing by single molecule counting. Angew. Chem. Int. Ed. 58, 3073–3076 (2019).
Delisle, B. P. et al. Thapsigargin selectively rescues the trafficking defective LQT2 channels G601S and F805C. J. Biol. Chem. 278, 35749–35754 (2003).
Magadán, J. G., Barbieri, M. A., Mesa, R., Stahl, P. D. & Mayorga, L. S. Rab22a regulates the sorting of transferrin to recycling endosomes. Mol. Cell. Biol. 26, 2595–2614 (2006).
van Galen, J. et al. Sphingomyelin homeostasis is required to form functional enzymatic domains at the trans-Golgi network. J. Cell Biol. 206, 609–618 (2014).
Modi, S., Halder, S., Nizak, C. & Krishnan, Y. Recombinant antibody mediated delivery of organelle-specific DNA pH sensors along endocytic pathways. Nanoscale 6, 1144–1152 (2014).
Delisle, B. P. et al. Intragenic suppression of trafficking-defective KCNH2 channels associated with long QT syndrome. Mol. Pharmacol. 68, 233–240 (2005).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Figure 1. 3D surface plots and selectivity studies. Figshare https://doi.org/10.6084/m9.figshare.23713359.v1 (2023).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Figure 2. 2-IM plots. Figshare https://doi.org/10.6084/m9.figshare.23713554.v1 (2023).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Figure 3. Colocalization data and 2-IM plots. Figshare https://doi.org/10.6084/m9.figshare.23713776.v1 (2023).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Figure 4. Colocalization data and 2-IM plots. Figshare https://doi.org/10.6084/m9.figshare.23713821.v1 (2023).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Figure 5. 2-IM plots. Figshare https://doi.org/10.6084/m9.figshare.23713833.v1 (2023).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Figure 6. 2-IM plots, I–V curves and peak current density plots. Figshare https://doi.org/10.6084/m9.figshare.23713851.v1 (2023).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Extended data figures. Fluorescence spectra, 2-IM plots, competition experiments, standard current–voltage (I–V) relationships. Figshare https://doi.org/10.6084/m9.figshare.23713899.v1 (2023).
Anees, P. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Supplementary figures. Fluorescence spectra, absorption spectra, 2-IM plots, colocalization data, blind test. Figshare https://doi.org/10.6084/m9.figshare.23713947.v1. (2023).
Acknowledgements
We thank E. Perozo and A. Lin Chun for valuable comments on the paper. We thank the integrated light microscopy facilities at the University of Chicago. Y.K. acknowledges funding from NIH grants DP1GM149751, 1R01NS112139-01A1, R21HL161825-01A1 (Y.K. and B.P.D), 1R01GM147197-01 and FA9550-19-0003 from the AFOSR, HFSP grant no. RGP0032/2022, and the Ono Pharma Foundation.
Author information
Authors and Affiliations
Contributions
P.A., A.S. and Y.K. designed the sensor and experiments related to its validation. P.A. designed and synthesized the TAC-Rh dye. P.A., B.P.D. and Y.K. designed experiments related to Kv11.1 channels. E.R.R. performed electrophysiology. P.A. performed all other experiments. P.A. and E.R.R. analyzed data. A.D. and A.B.M. provided TWIK2 KO BMDM cells. P.A., B.P.D. and Y.K. wrote the paper. All authors provided input on the paper.
Corresponding authors
Ethics declarations
Competing interests
Y.K. is co-founder of Esya Inc. and MacroLogic Inc., which use DNA nanodevices for diagnostics and therapeutics, respectively. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Haoxing Xu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 TAC-Rh fluorescence response as a function of both pH and K+.
a, Working principle of K+ sensing by TAC-Rh. b, Excitation (black) and emission (green) spectra of TAC-Rh. Emission intensity increased with increasing K+ concentration at pH = 7.0. c, Normalized O/R ratio of TAC-Rh/Alexa Fluor 647 with increasing [K+] at pH 7.0 and 6.0. Error bar represents mean + s.e.m. of three independent experiments. d, Fluorescence emission spectra of pHlicKerRE corresponding to TAC-Rh (green) and Alexa Fluor 647 (magenta) with increasing [K+] at pH = 7.0. e, Normalized O/R ratio of TAC-Rh/Alexa Fluor 647 with increasing [K+] at pH 7.0 Error bar represents mean + s.e.m. of three independent experiments.
Extended Data Fig. 2 Targeting modules (T) in organelle-specific pHlicKer variants.
a, pHlicKerRE localizes in recycling endosomes (REs) by transferrin receptor (TfR)-mediated endocytosis. T is an aptamer that binds TfR. b, pHlicKerTGN is retrogradely trafficked by an scFv-furin chimera to the trans Golgi network (TGN). T is a d(AT)4 sequence (cyan) in pHlicKer that sequence-specifically binds a single chain variable fragment (scFv) fused to the extracellular domain of furin. c, pHlicKerEE localizes in early endosomes (EEs) by scavenger receptor-mediated endocytosis. T is a duplex DNA domain, which is an excellent ligand for scavenger receptors. d–f, Three way (3W) junctions 3WRE, 3WTGN and 3WEE are made from the same sequences as pHlicKerRE, pHlicKerTGN and pHlicKerEE and lack the Alexa488 and TAC-Rh fluorophores for colocalization studies with fluorescent markers of the RE, TGN and EE. g, pHlicKerBiotin incorporates a biotin (grey pentagons) as indicated for immobilization on streptavidin coated beads.
Extended Data Fig. 3 Calibration of pHlicKer on beads.
a, Representative images of pHlicKerBiotin on beads clamped at indicated pH and K+ levels, imaged in the donor channel (D), acceptor channel (A), TMR (O), and Alexa Fluor 647 (R) channels. D/A and O/R are the corresponding pixel-wise pseudocolor images. (n = 100 beads). Scale bars, 5 μm. b–d, 2-IM profiles of beads clamped at indicated pH and [K+]. Experiments were performed in triplicate (n = 70–200 beads).
Extended Data Fig. 4 Targetability of 3WEE and 3WRE.
a, Uptake by HEK 293T cells expressing human scavenger receptor (hMSR1). Representative images of the uptake of Alexa 647 labelled 3WEE in untransfected and hMSR1 transfected HEK 293T cells. Scale bars, 5 μm. b, Normalized whole cell intensities for (a). Data represent mean ± s.e.m (n = 20–22 cells). hMSR1 expressing HEK 293T cells showed effective internalization of pHlicKerEE, revealing uptake is by scavenger receptors. c, Competition experiments with 3WRE and excess unlabelled transferrin (Tf) in HEK 293T cells. Representative fluorescence images of HEK 293T cells pulsed with 3WRE (500 nM) in the presence (+Tf, 20 μM) and absence (-Tf) of Tf. Cells are imaged in the Alexa 647 channel. AF, autofluorescence. Scale bars, 5 μm. d, Normalized intensities for (c). Data represent mean ± s.e.m (n = 18–22 cells). pHlicKerRE internalization by HEK 293T cell is competed out by excess Tf, revealing that uptake is mediated by transferrin receptor-mediated endocytosis.
Extended Data Fig. 5 Kv11.1 channel activity under transmembrane ion gradients equivalent to plasma membrane and recycling endosome.
a, b, Representative families of currents measured from cells stably expressing WT-Kv11.1 (black) or G601S- Kv11.1 (magenta) channel proteins using the voltage protocol shown in inset. a, Traces recorded using the standard extracellular saline or b, the modified extracellular saline to mimic recycling endosomes. Individual I-V relations were generated for each cell in each condition by plotting the peak current recorded during the test-pulse as a function of the pre-pulse (Fig. 6i,j). The individual I-V relations were described using a Boltzmann equation to calculate the IMAX (Fig. 6i–j), c, midpoint potential for IKv11.1 activation (V1/2), or d, the slope factor for IKv11.1 current activation (k). (Extended Data Fig. 5c: n = 9 for WT and n = 6 for G601S; Extended Data Fig. 5d: n = 4 for WT and n = 4 for G601S). Embedded box plots indicate the 25th–75th percentile. Boxes and bars represent the s.e.m. and standard deviation, respectively. *p=0.018; **p=0.001 (one-way ANOVA with Tukey post hoc test).
Supplementary information
Supplementary Information
Synthetic schemes 1–3, Supplementary Figs. 1–15, Notes 1–4 and Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Anees, P., Saminathan, A., Rozmus, E.R. et al. Detecting organelle-specific activity of potassium channels with a DNA nanodevice. Nat Biotechnol (2023). https://doi.org/10.1038/s41587-023-01928-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41587-023-01928-z
This article is cited by
-
Organellar electrophysiology: DNA nanodevices charging at the unmeasured
BMC Biology (2024)
-
DNA nanodevices map intracellular ions
Nature Biotechnology (2023)