Abstract
Signal peptides (SPs) are short amino acid sequences in the amino terminus of many newly synthesized proteins that target proteins into, or across, membranes. Bioinformatic tools can predict SPs from amino acid sequences, but most cannot distinguish between various types of signal peptides. We present a deep neural network-based approach that improves SP prediction across all domains of life and distinguishes between three types of prokaryotic SPs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Code availability
SignalP 5.0 is available at http://www.cbs.dtu.dk/services/SignalP/. The web version of SignalP 5.0 is free for all users, while the standalone package is free for academic users (and can be provided upon request) but is licensed for a fee to commercial users.
Data availability
The data sets used for training and testing SignalP 5.0 can be downloaded from http://www.cbs.dtu.dk/services/SignalP/data.php.
References
Nouwen, N., Berrelkamp, G. & Driessen, A. J. J. Mol. Biol. 372, 422–433 (2007).
Pohlschroder, M., Gimenez, M. I. & Jarrell, K. F. Curr. Opin. Microbiol. 8, 713–719 (2005).
Rapoport, T. A. Nature 450, 663–669 (2007).
Berks, B. C. Annu. Rev. Biochem. 84, 843–864 (2015).
von Heijne, G. Protein Eng. 2, 531–534 (1989).
Pohlschroder, M., Pfeiffer, F., Schulze, S. & Halim, M. F. A. FEMS Microbiol. Rev. 42, 694–717 (2018).
Sankaran, K. & Wu, H. C. J. Biol. Chem. 269, 19701–19706 (1994).
Szabo, Z. et al. J. Bacteriol. 189, 772–778 (2007).
Nielsen, H., Engelbrecht, J., Brunak, S. & von Heijne, G. Protein Eng. 10, 1–6 (1997).
Nielsen, H. & Krogh, A. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 122–130 (1998).
Bendtsen, J. D., Nielsen, H., von Heijne, G. & Brunak, S. J. Mol. Biol. 340, 783–795 (2004).
Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. Nat. Methods 8, 785–786 (2011).
Thompson, B. J. et al. Mol. Microbiol. 77, 943–957 (2010).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. Bioinformatics 28, 3150–3152 (2012).
Henikoff, S. & HenikoffJ. G. Proc. Natl Acad. Sci. USA 89, 10915–10919 (1992).
Frank, K. & Sippl, M. J. Bioinformatics 24, 2172–2176 (2008).
Altschul, S. F. et al. Nucleic Acids Res. 25, 3389–3402 (1997).
Matthews, B. W. Biochim. Biophys. Acta 405, 442–451 (1975).
Savojardo, C., Martelli, P. L., Fariselli, P. & Casadio, R. Bioinformatics 34, 1690–1696 (2017).
Bagos, P. G., Tsirigos, K. D., Plessas, S. K., Liakopoulos, T. D. & Hamodrakas, S. J. PEDS 22, 27–35 (2009).
Reynolds, S. M., Kall, L., Riffle, M. E., Bilmes, J. A. & Noble, W. S. PLoS Comput. Biol. 4, e1000213 (2008).
Kall, L., Krogh, A. & Sonnhammer, E. L. J. Mol. Biol. 338, 1027–1036 (2004).
Viklund, H., Bernsel, A., Skwark, M. & Elofsson, A. Bioinformatics 24, 2928–2929 (2008).
Tsirigos, K. D., Peters, C., Shu, N., Kall, L. & Elofsson, A. Nucleic Acids Res. 43, W401–W407 (2015).
Bagos, P. G., Nikolaou, E. P., Liakopoulos, T. D. & Tsirigos, K. D. Bioinformatics 26, 2811–2817 (2010).
Dilks, K., Rose, R. W., Hartmann, E. & Pohlschroder, M. J. Bacteriol. 185, 1478–1483 (2003).
UniProt Consortium. Nucleic Acids Res. 46, 2699 (2018).
Fraser, C. M. et al. Science 270, 397–403 (1995).
Sigrist, C. J. et al. Nucleic Acids Res. 41, D344–D347 (2013).
Bagos, P. G., Tsirigos, K. D., Liakopoulos, T. D. & Hamodrakas, S. J. J. Proteome. Res. 7, 5082–5093 (2008).
Dobson, L., Lango, T., Remenyi, I. & Tusnady, G. E. Nucleic Acids Res. 43, D283–D289 (2015).
Kozma, D., Simon, I. & Tusnady, G. E. Nucleic Acids Res. 41, D524–D529 (2013).
Juncker, A. S. et al. Protein Sci. 12, 1652–1662 (2003).
Kall, L., Krogh, A. & Sonnhammer, E. L. Bioinformatics 21, i251–i257 (2005).
Hiller, K., Grote, A., Scheer, M., Munch, R. & Jahn, D. Nucleic Acids Res. 32, W375–W379 (2004).
Gomi, M., Sonoyama, M. & Mitaku, S. Chem. Bio. Informat. J. 4, 142–147 (2004).
Bendtsen, J. D., Nielsen, H., Widdick, D., Palmer, T. & Brunak, S. BMC Bioinformatics 6, 167–173 (2005).
Zhang, Y. Z. & Shen, H. B. J. Chem. Inf. Model. 57, 988–999 (2017).
Chou, K. C. & Shen, H. B. Biochem. Biophys. Res. Commun. 357, 633–640 (2007).
Fariselli, P., Finocchiaro, G. & Casadio, R. Bioinformatics 19, 2498–2499 (2003).
LeCun, Y., Bengio, Y. & Hinton, G. Nature 521, 436–444 (2015).
Pan, S. J. & Yang, Q. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010).
Lafferty, J. D., McCallum, A. & Pereira, F. C. N. Proc. Eighteenth Int. Conf. Mach. Learn. 282–289 (2001).
Hochreiter, S. & Schmidhuber, J. Neural Comput. 9, 1735–1780 (1997).
Graves, A. Supervised sequence labelling. in Supervised Sequence Labelling with Recurrent Neural Networks 5–13, https://doi.org/10.1007/978-3-642-24797-2_2 (Springer, Berlin and Heidelberg, Germany, 2012).
Almagro Armenteros, J. J., Sonderby, C. K., Sonderby, S. K., Nielsen, H. & Winther, O. Bioinformatics 33, 3387–3395 (2017).
Zhou, J., & Troyanskaya, O. G. Proc. 31st Int. Conf. Mach. Learn. 753–745 (2014).
Bishop, C. Pattern Recognition and Machine Learning (Springer, New York, 2006).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Hutter, F., Hoos, H. H. & Leyton-Brown, K. Proc. 5th Int. Conf. Learn. Intell. Optimiz. 507–523 (2011).
Abadi, et al. Proc 12th USENIX Conf. Operat. Syst. Des. Implement. 265–283 (2016).
Acknowledgements
SB would like to acknowledge support from the Novo Nordisk Foundation (grant NNF14CC0001).
Author information
Authors and Affiliations
Contributions
J.J.A.A. designed the model architecture and trained the SignalP5 method with help from C.K.S. K.D.T. collected the training and test data sets, performed the benchmarks and analyzed results. C.K.S., T.N.P., O.W., S.B. and G.v.H. provided suggestions during the design of SignalP5. K.D.T and H.N wrote the paper with input from J.J.A.A., C.K.S. and O.W. H.N. supervized and guided the project. All authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The downloadable version of SignalP 5.0 has been commercialized by the Technical University of Denmark (it is licensed for a fee to commercial users). The revenue from these commercial sales is divided between the program developers (J.J.A.A., K.D.T., C.K.S., T.N.P., O.W., S.B., G.v.H. and H.N.) and the Technical University of Denmark.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Box plot of the probability of the predicted class for correct and incorrect predictions.
A probability close to 1 means a highly reliable prediction. For Archaea, Gram-Positive and Gram-Negative the probability threshold is 0.25, as there are four possible classes (Sec/SPI, Tat/SPI, Sec/SPII and Other). For Eukarya this threshold is 0.5, as it has only two classes (Sec/SPI and Other). A probability close to this threshold means a very unreliable prediction. All classes, namely Sec/SPI, Tat/SPI, Sec/SPII and Other are combined in this plot.
Supplementary Figure 2
Performance of SignalP 5.0 on cleavage site detection when considering a window of 0, 1, 2 and 3 amino acids centered on the real cleavage site.
Supplementary Figure 3
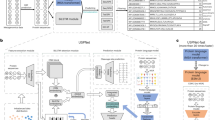
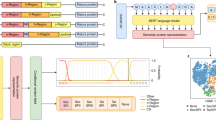
The SignalP 5.0 neural network architecture.
Supplementary information
Supplementary Information
Supplementary Figures 1–3, Supplementary Tables 1–12 and Supplementary Notes 1–3
Rights and permissions
About this article
Cite this article
Almagro Armenteros, J.J., Tsirigos, K.D., Sønderby, C.K. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37, 420–423 (2019). https://doi.org/10.1038/s41587-019-0036-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41587-019-0036-z
This article is cited by
-
Multi-Omics integration can be used to rescue metabolic information for some of the dark region of the Pseudomonas putida proteome
BMC Genomics (2024)
-
Aspartyl protease in the secretome of honey bee trypanosomatid parasite contributes to infection of bees
Parasites & Vectors (2024)
-
An apicoplast-localized deubiquitinase contributes to the cell growth and apicoplast homeostasis of Toxoplasma gondii
Veterinary Research (2024)
-
Transcriptomic profiling of different developmental stages reveals parasitic strategies of Wohlfahrtia magnifica, a myiasis-causing flesh fly
BMC Genomics (2024)
-
Expression of sex-specific molecular markers by Babesia bovis gametes
Parasites & Vectors (2024)