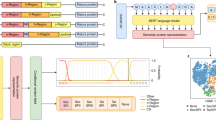
Abstract
Signal peptides (SPs) are essential to target and transfer transmembrane and secreted proteins to the correct positions. Many existing computational tools for predicting SPs disregard the extreme data imbalance problem and rely on additional group information of proteins. Here we introduce Unbiased Organism-agnostic Signal Peptide Network (USPNet), an SP classification and cleavage-site prediction deep learning method. Extensive experimental results show that USPNet substantially outperforms previous methods on classification performance by 10%. An SP-discovering pipeline with USPNet is designed to explore unprecedented SPs from metagenomic data. It reveals 347 SP candidates, with the lowest sequence identity between our candidates and the closest SP in the training dataset at only 13%. In addition, the template modeling scores between candidates and SPs in the training set are mostly above 0.8. The results showcase that USPNet has learnt the SP structure with raw amino acid sequences and the large protein language model, thereby enabling the discovery of unknown SPs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All the datasets we used, including training data, benchmark data, independent test data, proteome-wide study results and metagenomic study results are listed in Methods and are available at https://doi.org/10.17605/OSF.IO/NH3CF ref. 49. Source data are provided with this paper.
Code availability
The open-source codes of USPNet can be found at https://github.com/ml4bio/USPNet and the Code Ocean software capsule https://doi.org/10.24433/CO.8184163.v1 ref. 50.
References
von Heijne, G. Life and death of a signal peptide. Nature 396, 111–113 (1998).
Heijne, G. V. The signal peptide. J. Membr. Biol. 115, 195–201 (1990).
Bradshaw, N., Neher, S. B., Booth, D. S. & Walter, P. Signal sequences activate the catalytic switch of SRP RNA. Science 323, 127–130 (2009).
von Heijne, G. Patterns of amino acids near signal-sequence cleavage sites. Eur. J. Biochem. 133, 17–21 (1983).
Madani, A. et al. Large language models generate functional protein sequences across diverse families. Nat. Biotechnol. 41, 1099–1106 (2023).
Frank, K. & Sippl, M. J. High-performance signal peptide prediction based on sequence alignment techniques. Bioinformatics 24, 2172–2176 (2008).
Petersen, T. N., Brunak, S., Von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 (2011).
Savojardo, C., Martelli, P. L., Fariselli, P. & Casadio, R. DeepSig: deep learning improves signal peptide detection in proteins. Bioinformatics 10, 1690–1696 (2017).
Armenteros, J. J. A. et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37, 420–423 (2019).
Teufel, F. et al. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025 (2022).
Juncker, A. S. et al. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662 (2003).
Bagos, P. G., Tsirigos, K. D., Liakopoulos, T. D. & Hamodrakas, S. J. Prediction of lipoprotein signal peptides in Gram-positive bacteria with a hidden Markov model. J. Proteome Res. 7, 5082–5093 (2008).
Bendtsen, J. D., Nielsen, H., Widdick, D., Palmer, T. & Brunak, S. Prediction of twin-arginine signal peptides. BMC Bioinformatics 6, 167 (2005).
Pasolli, E. et al. Accessible, curated metagenomic data through experimenthub. Nat. Methods 14, 1023–1024 (2017).
Sczyrba, A. et al. Critical assessment of metagenome interpretation—a benchmark of metagenomics software. Nat. Methods 14, 1063–1071 (2017).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl Acad. Sci. USA 118, e2016239118 (2021).
Rao, R. M. et al. MSA transformer. In Proc. 38th International Conference on Machine Learning, Proc. Machine Learning Research Vol. 139 (eds Meila, M. & Zhang, T.) 8844–8856 (PMLR, 2021).
Biswas, S., Khimulya, G., Alley, E. C., Esvelt, K. M. & Church, G. M. Low-N protein engineering with data-efficient deep learning. Nat. Methods 18, 389–396 (2021).
Thireou, T. & Reczko, M. Bidirectional long short-term memory networks for predicting the subcellular localization of eukaryotic proteins. IEEE/ACM Trans. Comput. Biol. Bioinform. 4, 441–446 (2007).
Cao, K., Wei, C., Gaidon, A., Arechiga, N. & Ma, T. Learning imbalanced datasets with label-distribution-aware margin loss. Adv. Neural Inf. Process. Syst. 32, 1567–1578 (2019).
Mnih, V. et al. Recurrent models of visual attention. Adv. Neural Inf. Process. Syst. 27, 2204–2212 (2014).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Lin, T.-Y., Goyal, P., Girshick, R., He, K. & Dollár, P. Focal loss for dense object detection. In Proc. IEEE International Conference on Computer Vision 2980–2988 (IEEE, 2017).
Elnaggar, A. et al. ProtTrans: toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2021).
Armenteros, J. J. A. et al. Detecting sequence signals in targeting peptides using deep learning. Life Sci. Alliance 2, e201900429 (2019).
Ma, Y. et al. Identification of antimicrobial peptides from the human gut microbiome using deep learning. Na. Biotechnol. 40, 921–931 (2022).
Han, S. et al. Novel signal peptides improve the secretion of recombinant Staphylococcus aureus alpha toxinH35L in Escherichia coli. AMB Express 7, 93 (2017).
Consortium, T. U. UniProt: the universal protein knowledgebase in 2023. Nucleic Acids Res. 51, D523–D531 (2022).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Consortium, U. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2019).
Sigrist, C. J. et al. New and continuing developments at prosite. Nucleic Acids Res. 41, D344–D347 (2012).
Dobson, L., Lango, T., Reményi, I. & Tusnády, G. E. Expediting topology data gathering for the TOPDB database. Nucleic Acids Res. 43, D283–D289 (2015).
Gíslason, M. H., Nielsen, H., Armenteros, J. J. A. & Johansen, A. R. Prediction of GPI-anchored proteins with pointer neural networks. Curr. Res. Biotechnol. 3, 6–13 (2021).
Li, W. & Godzik, A. CD-HIT: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Youngblut, N. D. et al. Large-scale metagenome assembly reveals novel animal-associated microbial genomes, biosynthetic gene clusters, and other genetic diversity. mSystems 5, e01045-20 (2020).
Looft, T., Bayles, D., Alt, D. & Stanton, T. Complete genome sequence of Coriobacteriaceae strain 68-1-3, a novel mucus-degrading isolate from the swine intestinal tract. Genome Announc. 3, e01143-15 (2015).
Zhou, S. et al. Characterization of metagenome-assembled genomes and carbohydrate-degrading genes in the gut microbiota of Tibetan pig. Front. Microbiol. 11, 595066 (2020).
Chen, C. et al. Prevotella copri increases fat accumulation in pigs fed with formula diets. Microbiome 9, 175 (2021).
Groussin, M. et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell 184, 2053–2067 (2021).
Tilocca, B. et al. Dietary changes in nutritional studies shape the structural and functional composition of the pigs’ fecal microbiome—from days to weeks. Microbiome 5, 144 (2017).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Steinegger, M., Mirdita, M. & Söding, J. Protein-level assembly increases protein sequence recovery from metagenomic samples manyfold. Nat. Methods 16, 603–606 (2019).
Mirdita, M. et al. UniCclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 45, D170–D176 (2017).
Steinegger, M. et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinformatics 20, 473 (2019).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at arXiv https://doi.org/10.48550/arXiv.1802.03426 (2018).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
DeLano, W. L. et al. PyMOL: an open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 40, 82–92 (2002).
Zhang, Y. & Skolnick, J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 (2005).
Shen, J. et al. Unbiased organism-agnostic and highly sensitive signal peptide predictor with deep protein language model. OSF https://doi.org/10.17605/OSF.IO/NH3CF (2023).
Shen, J. et al. Unbiased organism-agnostic and highly sensitive signal peptide predictor with deep protein language model. Code Ocean https://doi.org/10.24433/CO.8184163.v1 (2023).
Acknowledgements
Special thanks to the people who suggested that we evaluate models on the 40% cut-off benchmark set. The work described in this paper was supported by a grant from the Research Grants Council of the Hong Kong Special Administrative Region, China (project number CUHK 24204023, to Y.L.) and a grant from Innovation and Technology Commission of the Hong Kong Special Administrative Region, China (project number GHP/065/21SZ, to Y.L.). The work was partially supported by the National Key R&D Program of China (NO.2022ZD0160101).
Author information
Authors and Affiliations
Contributions
Y.L., J.S. and S.C. designed the computational method. J.S., Q.Y. and S.C. implemented the main algorithm. J.S., Q.Y., S.C., Q.T. and J.L. did the experiments. J.S. and Q.Y. performed the analysis. J.S., Q.Y. and S.C. wrote the paper. Y.L. supervised the project. All authors read and approved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Computational Science thanks Rita Casadio and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Jie Pan, in collaboration with the Nature Computational Science team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–7, Supplementary Figs. 1–6 and Tables 1–21.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shen, J., Yu, Q., Chen, S. et al. Unbiased organism-agnostic and highly sensitive signal peptide predictor with deep protein language model. Nat Comput Sci 4, 29–42 (2024). https://doi.org/10.1038/s43588-023-00576-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43588-023-00576-2