Abstract
Transient receptor potential (TRP) channels are a large, eukaryotic ion channel superfamily that control diverse physiological functions, and therefore are attractive drug targets1,2,3,4,5. More than 210 structures from more than 20 different TRP channels have been determined, and all are tetramers4. Despite this wealth of structures, many aspects concerning TRPV channels remain poorly understood, including the pore-dilation phenomenon, whereby prolonged activation leads to increased conductance, permeability to large ions and loss of rectification6,7. Here, we used high-speed atomic force microscopy (HS-AFM) to analyse membrane-embedded TRPV3 at the single-molecule level and discovered a pentameric state. HS-AFM dynamic imaging revealed transience and reversibility of the pentamer in dynamic equilibrium with the canonical tetramer through membrane diffusive protomer exchange. The pentamer population increased upon diphenylboronic anhydride (DPBA) addition, an agonist that has been shown to induce TRPV3 pore dilation. On the basis of these findings, we designed a protein production and data analysis pipeline that resulted in a cryogenic-electron microscopy structure of the TRPV3 pentamer, showing an enlarged pore compared to the tetramer. The slow kinetics to enter and exit the pentameric state, the increased pentamer formation upon DPBA addition and the enlarged pore indicate that the pentamer represents the structural correlate of pore dilation. We thus show membrane diffusive protomer exchange as an additional mechanism for structural changes and conformational variability. Overall, we provide structural evidence for a non-canonical pentameric TRP-channel assembly, laying the foundation for new directions in TRP channel research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data and materials to draw the conclusions in this paper are presented in the main text, figures and the extended data figures and supplementary videos. The cryo-EM maps of the TRPV3 tetramer and pentamer have been deposited in the Electron Microscopy Data Bank with accession codes EMD-40181 and EMD-40183, respectively, and their structural models have been deposited in the PDB with accession codes 8GKA and 8GKG, respectively (Extended Data Table 1). Further data can be received from the corresponding author upon reasonable request.
References
Peng, G., Shi, X. & Kadowaki, T. Evolution of TRP channels inferred by their classification in diverse animal species. Mol. Phylogenet. Evol. 84, 145–157 (2015).
Himmel, N. J. & Cox, D. N. Transient receptor potential channels: current perspectives on evolution. Proc. R. Soc. B. Biol. Sci. 287, 20201309 (2020).
Khalil, M. et al. Functional role of transient receptor potential channels in immune cells and epithelia. Front. Immunol. 9, 174 (2018).
Huffer, K. E., Aleksandrova, A. A., Jara-Oseguera, A., Forrest, L. R. & Swartz, K. J. Global alignment and assessment of trp channel transmembrane domain structures to explore functional mechanisms. eLife 9, e58660 (2020).
Moran, M. M. TRP channels as potential drug targets. Annu. Rev. Pharmacol. Toxicol. 58, 309–330 (2018).
Ferreira, L. G. B. & Faria, R. X. TRPing on the pore phenomenon: what do we know about transient receptor potential ion channel-related pore dilation up to now? J. Bioenerg. Biomembr. 48, 1–12 (2016).
Zheng, J. & Ma, L. Structure and function of the ThermoTRP channel pore. Curr. Top. Membr. 74, 233–257 (2014).
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013).
Bai, X. C., Fernandez, I. S., McMullan, G. & Scheres, S. H. W. Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. eLife 2013, 2–13 (2013).
Caterina, M. J. et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824 (1997).
Kashio, M. & Tominaga, M. TRP channels in thermosensation. Curr. Opin. Neurobiol. 75, 102591 (2022).
van Goor, M. K. C., Hoenderop, J. G. J. & van der Wijst, J. TRP channels in calcium homeostasis: from hormonal control to structure-function relationship of TRPV5 and TRPV6. Biochim. Biophys. Acta Mol. Cell Res. 1864, 883–893 (2017).
Pumroy, R. A. et al. Molecular mechanism of TRPV2 channel modulation by cannabidiol. eLife 8, e48792 (2019).
Xu, H., Delling, M., Jun, J. C. & Clapham, D. E. Oregano, thyme and clove-derived flavors and skin sensitizers activate specific TRP channels. Nat. Neurosci. 9, 628–635 (2006).
Deng, Z. et al. Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat. Struct. Mol. Biol. 25, 252–260 (2018).
Deng, Z. et al. Gating of human TRPV3 in a lipid bilayer. Nat. Struct. Mol. Biol. 27, 635–644 (2020).
Nadezhdin, K. D. et al. Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel. Nat. Struct. Mol. Biol. 28, 564–572 (2021).
Zubcevic, L. et al. Conformational ensemble of the human TRPV3 ion channel. Nat. Commun. 9, 4773 (2018).
Nilius, B., Bíró, T. & Owsianik, G. TRPV3: time to decipher a poorly understood family member! J. Physiol. 592, 295–304 (2014).
Bautista, D. & Julius, D. Fire in the hole: pore dilation of the capsaicin receptor TRPV1. Nat. Neurosci. 11, 528–529 (2008).
Chung, M. K., Güler, A. D. & Caterina, M. J. TRPV1 shows dynamic ionic selectivity during agonist stimulation. Nat. Neurosci. 11, 555–564 (2008).
Zhang, K., Julius, D. & Cheng, Y. Structural snapshots of TRPV1 reveal mechanism of polymodal functionality. Cell 184, 5138–5150.e12 (2021).
Canul-Sánchez, J. A. et al. Different agonists induce distinct single-channel conductance states in TRPV1 channels. J. Gen. Physiol. 150, 1735–1746 (2018).
Nieto-Posadas, A. et al. Lysophosphatidic acid directly activates TRPV1 through a C-terminal binding site. Nat. Chem. Biol. 8, 78–85 (2012).
Chung, M. K., Güler, A. D. & Caterina, M. J. Biphasic currents evoked by chemical or thermal activation of the heat-gated ion channel, TRPV3. J. Biol. Chem. 280, 15928–15941 (2005).
Chen, J. et al. Pore dilation occurs in TRPA1 but not in TRPM8 channels. Mol. Pain 5, 2–7 (2009).
Banke, T. G., Chaplan, S. R. & Wickenden, A. D. Dynamic changes in the TRPA1 selectivity filter lead to progressive but reversible pore dilation. Am. J. Physiol. Cell Physiol. 298, 1457–1468 (2010).
Zubcevic, L., Le, S., Yang, H. & Lee, S. Y. Conformational plasticity in the selectivity filter of the TRPV2 ion channel. Nat. Struct. Mol. Biol. 25, 405–415 (2018).
Uchihashi, T. & Scheuring, S. Applications of high-speed atomic force microscopy to real-time visualization of dynamic biomolecular processes. Biochim. Biophys. Acta Gen. Subj. 1862, 229–240 (2018).
Heath, G. R. & Scheuring, S. Advances in high-speed atomic force microscopy (HS-AFM) reveal dynamics of transmembrane channels and transporters. Curr. Opin. Struct. Biol. 57, 93–102 (2019).
Misetic, V., Reiners, O., Krauss, U. & Jaeger, K.-E. NanoDSF thermal unfolding analysis of proteins without tryptophan residues (Application Note NT‐PR‐007). NanoTemperTech https://resources.nanotempertech.com/application-notes/application-note-nt-pr-007-unfolding-without-tryptophan (2016).
Real-Hohn, A., Groznica, M., Löffler, N., Blaas, D. & Kowalski, H. nanoDSF: in vitro label-free method to monitor picornavirus uncoating and test compounds affecting particle stability. Front. Microbiol. 11, 1442 (2020).
Grubisha, O. et al. Pharmacological profiling of the TRPV3 channel in recombinant and native assays. Br. J. Pharmacol. 171, 2631–2644 (2014).
Yu, F. H., Yarov-Yarovoy, V., Gutman, G. A. & Catterall, W. A. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol. Rev. 57, 387–395 (2005).
Nadezhdin, K. D. et al. Extracellular cap domain is an essential component of the TRPV1 gating mechanism. Nat. Commun. 12, 4–11 (2021).
Singh, A. K., Saotome, K. & Sobolevsky, A. I. Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci. Rep. 7, 10669 (2017).
Yelshanskaya, M. V. & Sobolevsky, A. I. Ligand-binding sites in vanilloid-subtype TRP channels. Front. Pharmacol. 13, 900623 (2022).
Singh, A. K., McGoldrick, L. L. & Sobolevsky, A. I. Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat. Struct. Mol. Biol. 25, 805–813 (2018).
Zubcevic, L., Borschel, W. F., Hsu, A. L., Borgnia, M. J. & Lee, S. Y. Regulatory switch at the cytoplasmic interface controls trpv channel gating. eLife 8, e47746 (2019).
Ni, C. et al. A novel mutation in TRPV3 gene causes atypical familial Olmsted syndrome. Sci. Rep. 6, 21815 (2016).
Duchatelet, S. et al. A new TRPV3 missense mutation in a patient with Olmsted syndrome and erythromelalgia. JAMA Dermatol. 150, 303–306 (2014).
Jiang, Y. et al. Membrane-mediated protein interactions drive membrane protein organization. Nat. Commun. 13, 7373 (2022).
Hazan, A., Kumar, R., Matzner, H. & Priel, A. The pain receptor TRPV1 displays agonist-dependent activation stoichiometry. Sci. Rep. 5, 12278 (2015).
Sente, A. et al. Differential assembly diversifies GABAA receptor structures and signalling. Nature 604, 190–194 (2022).
Noviello, C. M., Kreye, J., Teng, J., Prüss, H. & Hibbs, R. E. Structural mechanisms of GABAA receptor autoimmune encephalitis. Cell 185, 2469–2477 (2022).
Cheng, W., Yang, F., Takanishi, C. L. & Zheng, J. Thermosensitive TRPV channel subunits coassemble into heteromeric channels with intermediate conductance and gating properties. J. Gen. Physiol. 129, 191–207 (2007).
Bleakman, D., Broroson, J. R. & Miller, R. J. The effects of capsaicin on voltage-gated calcium currents and calcium signals in cultured dorsal root ganglion cells. Br. J. Pharmacol. 101, 423–431 (1990).
Evans, A. R., Nicol, G. D. & Vasko, M. R. Differential regulation of evoked peptide release by voltage-sensitive calcium channels in rat sensory neurons. Brain Res. 712, 265–273 (1996).
Jancso, G. Pathobiological reactions of C‐fibre primary sensory neurones to peripheral nerve injury. Exp. Physiol. 77, 405–431 (1992).
Goehring, A. et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9, 2574–2585 (2014).
Sanganna Gari, R. R. et al. Correlation of membrane protein conformational and functional dynamics. Nat. Commun. 12, 4363 (2021).
Heath, G. R. et al. Localization atomic force microscopy. Nature 594, 385–390 (2021).
Matin, T. R., Heath, G. R., Scheuring, S. & Boudker, O. Millisecond dynamics of an unlabeled amino acid transporter. Nat. Commun. 11, 5016 (2020).
Rangl, M., Schmandt, N., Perozo, E. & Scheuring, S. Real time dynamics of gating-related conformational changesin CorA. eLife 8, e47322 (2019).
Lin, Y. C. et al. Force-induced conformational changes in PIEZO1. Nature 573, 230–234 (2019).
Ruan, Y. et al. Structural titration of receptor ion channel GLIC gating by HS-AFM. Proc. Natl Acad. Sci. USA 115, 10333–10338 (2018).
Ruan, Y. et al. Direct visualization of glutamate transporter elevator mechanism by high-speed AFM. Proc. Natl Acad. Sci. USA 114, 1584–1588 (2017).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Oh, S. et al. Differential ion dehydration energetics explains selectivity in the non-canonical lysosomal K+ channel TMEM175. eLife 11, e75122 (2022).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J 478, 4169–4185 (2021).
Terwilliger, T. C., Sobolev, O. V., Afonine, P. V., Adams, P. D. & Read, R. J. Density modification of cryo-EM maps. Acta Crystallogr. Sect. D Struct. Biol. 76, 912–925 (2020).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Zwart, P. H. et al. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 (2008).
Emsley, P. & Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D: Biol. Crystallogr. 60, 2126–2132 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D: Biol. Crystallogr. 66, 12–21 (2010).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. Sect. D Struct. Biol. 74, 531–544 (2018).
Acknowledgements
We thank A. Accardi and J. Dittman for important discussions. Negative-stain EM data were collected in the Electron Microscopy & Histology services of the Weill Cornell Medicine Microscopy & Image Analysis Core using a transmission electron microscope purchased with funds from an National Institutes of Health (NIH) Shared Instrumentation grant (no. S10RR027699) for Shared Resources. Cryo-EM data were collected at the Simons Electron Microscopy Center at the New York Structural Biology Center, with support from the Simons Foundation (grant no. SF349247). Work in the Scheuring laboratory is partly supported by grants from the NIH, National Center for Complementary and Integrative Health (NCCIH), grant no. DP1AT010874 (to S.S.) and National Institute of Neurological Disorders and Stroke (NINDS), grant no. R01NS110790 (to S.S.). Work in the Yuan laboratory is partly supported by grant no. NIH NINDS R01NS099341 (to P.Y.). Work in the Nimigean laboratory is partly supported by grant no. NIH NIMGS R01 GM088352 (to C.M.N.) and grant no. NIH NIMGS F32 GM145091 (to E.D.K.). S.L. is an awardee of the Weizmann Institute of Science Women’s Postdoctoral Career Development Award.
Author information
Authors and Affiliations
Contributions
S.L. and S.S. designed the study. J.Z. and P.Y. expressed and purified protein from P. pastoris cells. S.L., J.M.B. and E.D.K. expressed and purified protein from HEK GnTI− cells. S.L. and J.M.B. reconstituted protein. S.L. performed negative-stain EM imaging. S.L., E.D.K. and C.M.N. performed and analysed electrophysiology measurements. S.L. and J.M.B. performed HS-AFM imaging. S.L. and Y.J. performed HS-AFM data analysis. S.L. and J.M.B. performed nanoDSF experiments and analysis. S.L. and J.M.B. performed cryo-EM sample preparation and data collection. S.L., E.D.K. and N.P. analysed single-particle cryo-EM data. S.L. and S.S. performed channel structure analysis. Y.J. performed and analysed oligomer simulation. S.L. and S.S. wrote the paper. All authors edited the manuscript. S.S. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Ute Hellmich, Thomas Voets and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
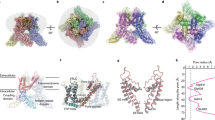
Extended Data Fig. 1 TRP-channel structures.
Representative structures (out of >210 structures) of the 23 TRP-channels solved so far. All structures are tetramers, with the four subunits coloured in wheat, green, purple and yellow. Each structure is depicted in surface representation, shown from the intracellular (top) and side (bottom) views. The question mark in the crTRP1 panel signifies that the subfamily to which crTRP1 belongs is yet unknown.
Extended Data Fig. 2 Single-channel recordings of TRPV3.
(a) and (b) Representative single-channel recordings of TRPV3 in the absence (a) and presence (b) of 100 μM DPBA, at −50 and 50 mV. (c) Since-channel current-voltage (IV) curves of TRPV3 obtained from −100 to 100 mV, in the absence and presence of 100 μM DPBA. (d) Single-channel open probabilities determined from recordings obtained at −50 mV, in the absence and presence of 100 μM DPBA. Open probability values (0.27 ± 0.01 and 0.78 ± 0.05 respectively) were derived as the mean values +/− s.e.m. from n ≥ 3 independent experiments (circles). Statistical significance was assessed with the one-tailed Welch’s T-test, yielding a significant (p-value = 0.0007) increase in open channel probability following DPBA addition. All recordings were performed on TRPV3 channels from one purification following the same protein expression and purification protocol as for the cryo-EM analysis, and reconstituted following the same protocol as for the HS-AFM analysis though at higher lipid-to-protein ratio (LPR between 5 and 20 for electrophysiology recordings vs. LPR between 0.5 and 2.5 for HS-AFM experiments). *** p-value < 0.005.
Extended Data Fig. 3 The TRPV3 tetramer and pentamer are reversible.
(a) to (f) Tetramer to pentamer transitions. (g) to (k) Pentamer to tetramer transitions. (l) Tetramer-pentamer-tetramer transition. (m) Pentamer-tetramer-pentamer-tetramer-pentamer-tetramer transition. White arrowheads indicate the occasionally observed monomers ‘attacking’ and inserting into tetramers to yield pentamers, and the observed monomers dissociating from pentamers to yield tetramers.
Extended Data Fig. 4 TRPV3 tetramers can breakup into fragments and reform.
(a) TRPV3 tetramers and pentamers coexist alongside TRPV3 ≤ 3 protomer fragments. Grey arrowheads indicate monomer (1), dimer (2), and trimer (3) fragments. (b) and (c) TRPV3 tetramer breakups into trimer and dimer. (d) TRPV3 fragments form a stable tetramer, which then breaks apart.
Extended Data Fig. 5 Workflow for cryo-EM reconstruction of the TRPV3 tetramer and pentamer.
Flowchart for the cryo-EM data processing, particle picking, classification, and reconstruction, enabling map reconstruction of the tetramer at 2.55Å resolution and for the pentamer at 4.38Å resolution. Unless otherwise stated, all processing steps were conducted in cryoSPARC version 3.3.2. Dashed lines indicate the inputs used for the iterative cycles of heterogenous refinement.
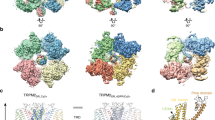
Extended Data Fig. 6 Cryo-EM density maps of the TRPV3 tetramer and pentamer.
(a) and (b) Cryo-EM reconstructed maps of the TRPV3 tetramer (a) and pentamer (b), colored according to local resolution using a rainbow colour scale. (c) and (d) Representative cryo-EM densities of the tetramer (contour level at 5.5 RMSD) (c) and pentamer (contour level at 4.16 RMSD) (d), at 2.55Å and 4.38Å resolution, respectively (the TMDs in the pentamer map are of ~5.0-5.5 Å resolution, see local resolution color scheme in (b)).
Extended Data Fig. 7 Structural comparison of the TRPV3 tetramer and pentamer cryo-EM structures.
(a) and (b) TRPV3 tetramer (a) and pentamer (b) structures, colored according to domains: ARD in purple, VSLD in yellow, PD in pink, SF in green, TRP helix in wheat, coupling domain in light blue. (c) to (e) Superposition of a pentamer subunit (purple) onto the tetramer subunit (green), aligned with respect to the PD, indicating a hinge-motion in the pentamer monomer by 18º, as manifested by rotation of the ARD (c), VSLD, and TRP helix (d). This hinge-motion enables preservation of the inter-subunit interactions of S5 with S1 and S4, and the SF-SF and S6-S6 interactions (e). Neighboring subunits in gray. Open book graphics of the tetramer (f, green) and pentamer (g, purple) inter-subunit contact areas. Contact areas are colored in pink (in the tetramer) and yellow (in the pentamer).
Extended Data Fig. 8 Model of the reversible transition between the tetrameric and pentameric TRPV3 states.
TRPV3 tetramers may dissociate. Dissociation is favored by activation, due to destabilization of the interprotomer interaction, notably the VSLD-PD domain-swap interface, by helix-intercalating molecules (e.g., capsaicin or LPA in TRPV1, DPBA in TRPV3, temperature). Monomers may ‘attack’ and insert into tetramers to yield pentamers with an estimated ~2.4-fold enlarged pore diameter at the SF. Pentamers, lifetime of ~3 min, are less stable than tetramers due to more fragile VSLD-PD interfaces, and shed subunits to regain the tetrameric state (figure created with BioRender.com).
Extended Data Fig. 9 Simulation of oligomeric state transitions.
(a) Visualization of simulated traces. Each column represents an independent space that can be either empty (0) or occupied by a molecule with a specific oligomeric state (1, 2, 3, 4 or 5). Total space: 5250. Total time step: 2000. Initial setup (time step = 1): 5000 out of 5250 spaces had a value of 4 (tetramers) and 250 out of the 5250 spaces had a value of 0 (empty). (b) and (c) Close-up views of the simulated traces in (a) as indicated by the dashed boxes. 1: Empty → monomer → dimer → trimer → tetramer → pentamer transition. 2: Transitions between tetramer and pentamer states (with short pentamer dwell-times). 3: A long pentamer state event. 4: pentamer → tetramer → trimer → dimer → monomer → empty transition. (d) Time-evolution of oligomer counts. Top panel: Tetramers. Middle panel: Pentamers and lower oligomers (trimer, dimer, and monomer) aggregated. Bottom panel: Trimers, dimers and monomers. (e) Oligomer state dwell-times. Left to right: Lower oligomers (n = 34984), tetramer (n = 278141), and pentamer (n = 331579).
Supplementary information
Supplementary Information
This file contains Supplementary Figs. 1–6, Tables 1–3 and Code.
Supplementary Video 1
Overview HS-AFM video of TRPV3 reconstitution. Frame rate, 1 s per frame. Pixel sampling, 0.80 nm per pixel.
Supplementary Video 2
Overview HS-AFM video of TRPV3 reconstitution. Frame rate, 2 s per frame. Pixel sampling, 0.80 nm per pixel.
Supplementary Video 3
Overview HS-AFM video of TRPV3 reconstitution. Frame rate, 1 s per frame. Pixel sampling, 0.27 nm per pixel.
Supplementary Video 4
Overview HS-AFM video of TRPV3 reconstitution. Frame rate, 1 s per frame. Pixel sampling, 0.40 nm per pixel.
Supplementary Video 5
Overview HS-AFM video of TRPV3 reconstitution revealing several channels with pentameric oligomeric states. Frame rate, 1.5 s per frame. Pixel sampling, 0.33 nm per pixel.
Supplementary Video 6
Overview HS-AFM video of TRPV3 reconstitution revealing several channels with pentameric oligomeric states. Frame rate, 1 s per frame. Pixel sampling, 0.50 nm per pixel.
Supplementary Video 7
Overview HS-AFM video of TRPV3 reconstitution revealing several channels with pentameric oligomeric states. Frame rate, 1 s per frame. Pixel sampling, 0.40 nm per pixel.
Supplementary Video 8
Overview HS-AFM video of TRPV3 reconstitution revealing several channels with pentameric oligomeric states. Frame rate, 0.5 s per frame. Pixel sampling, 0.48 nm per pixel.
Supplementary Video 9
Overview HS-AFM video of TRPV3 reconstitution revealing several channels with pentameric oligomeric states. Frame rate, 2 s per frame. Pixel sampling, 0.40 nm per pixel.
Supplementary Video 10
High-resolution HS-AFM videos of tetrameric TRPV3 channels. Frame rate, 1 s per frame. Pixel sampling, 0.20 nm per pixel.
Supplementary Video 11
High-resolution HS-AFM videos of a tetrameric and pentameric TRPV3 channel. Frame rate, 0.3 s per frame. Pixel sampling, 0.12 nm per pixel.
Supplementary Video 12
High-resolution HS-AFM videos of pentameric TRPV3 channels. Frame rate, 1.5 s per frame. Pixel sampling, 0.25 nm per pixel.
Supplementary Video 13
High-resolution HS-AFM videos of a tetrameric and pentameric TRPV3 channel. Frame rate, 1 s per frame. Pixel sampling, 0.35 nm per pixel.
Supplementary Video 14
High-resolution HS-AFM videos of a tetrameric and pentameric TRPV3 channel. Frame rate, 1 s per frame. Pixel sampling, 0.20 nm per pixel.
Supplementary Video 15
HS-AFM video of TRPV3 reconstitution revealing a tetramer–pentamer transition. Left panel shows an overview of TRPV3 reconstitution. Top right panel shows single-molecule tetramer–pentamer transition. Bottom right panel shows the average (t,t + 2) of single-molecule tetramer–pentamer transition. Frame rate, 2 s per frame. Pixel sampling, 0.28 nm per pixel.
Supplementary Video 16
HS-AFM video of TRPV3 reconstitution revealing a tetramer–pentamer transition. Left panel shows an overview of TRPV3 reconstitution. Top right panel shows single-molecule tetramer–pentamer transition. Bottom right panel shows the average (t,t + 2) of single-molecule tetramer–pentamer transition. Frame rate, 2 s per frame. Pixel sampling, 0.67 nm per pixel.
Supplementary Video 17
HS-AFM video of TRPV3 reconstitution revealing a tetramer–pentamer transition. Left panel shows an overview of TRPV3 reconstitution. Top right panel shows single-molecule tetramer–pentamer transition. Bottom right panel shows the average (t,t + 2) of single-molecule tetramer–pentamer transition. Frame rate, 1 s per frame. Pixel sampling, 0.32 nm per pixel.
Supplementary Video 18
HS-AFM video of TRPV3 reconstitution revealing two pentamer–tetramer transitions. Left panel shows an overview of TRPV3 reconstitution. Top right panel shows single-molecule pentamer–tetramer transitions. Bottom right panel shows the verage (t,t + 2) of single-molecule pentamer–tetramer transitions. Frame rate, 1 s per frame. Pixel sampling, 0.25 nm per pixel.
Supplementary Video 19
HS-AFM video of TRPV3 reconstitution revealing a pentamer–tetramer transitions. Left panel shows an overview of TRPV3 reconstitution. Top right panel shows the single-molecule pentamer–tetramer transition. Bottom right panel shows the average (t,t + 2) of single-molecule pentamer–tetramer transition. Frame rate, 1 s per frame. Pixel sampling, 0.35 nm per pixel.
Supplementary Video 20
HS-AFM video of TRPV3 reconstitution revealing a pentamer–tetramer transitions. Left panel shows an overview of TRPV3 reconstitution. Top right panel shows the single-molecule pentamer–tetramer transition. Bottom right panel shows the average (t,t + 2) of single-molecule pentamer–tetramer transition. Frame rate, 1 s per frame. Pixel sampling, 0.32 nm per pixel.
Supplementary Video 21
HS-AFM video of TRPV3 reconstitution revealing a complete tetramer–pentamer–tetramer transitions. Left panel shows an overview of TRPV3 reconstitution. Top right panel shows the single-molecule complete tetramer–pentamer–tetramer transition. Bottom right panel shows the average (t,t + 2) of single-molecule complete tetramer–pentamer–tetramer transition. Frame rate, 1.5 s per frame. Pixel sampling, 0.33 nm per pixel.
Supplementary Video 22
Overview HS-AFM video of TRPV3 reconstitution in the presence of 320 μM DPBA, revealing several channels with pentameric oligomeric states. Frame rate, 1 s per frame. Pixel sampling, 0.40 nm per pixel.
Supplementary Video 23
Overview HS-AFM video of TRPV3 reconstitution in the presence of 320 μM DPBA, revealing several channels with pentameric oligomeric states. Frame rate, 2 s per frame. Pixel sampling, 0.67 nm per pixel.
Supplementary Video 24
Overview HS-AFM video of TRPV3 reconstitution in the presence of 320 μM DPBA, revealing several channels with pentameric oligomeric states. Frame rate, 1 s per frame. Pixel sampling, 0.33 nm per pixel.
Supplementary Video 25
Overview HS-AFM video of TRPV3 reconstitution in the presence of 320 μM DPBA, revealing several channels with pentameric oligomeric states. Frame rate, 1 s per frame. Pixel sampling, 0.40 nm per pixel.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lansky, S., Betancourt, J.M., Zhang, J. et al. A pentameric TRPV3 channel with a dilated pore. Nature 621, 206–214 (2023). https://doi.org/10.1038/s41586-023-06470-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-06470-1
This article is cited by
-
A small-molecule activation mechanism that directly opens the KCNQ2 channel
Nature Chemical Biology (2024)
-
Structural heterogeneity of the ion and lipid channel TMEM16F
Nature Communications (2024)
-
Dynamic ion channel defies dogma
Nature (2023)
-
Intrinsically disordered regions in TRPV2 mediate protein-protein interactions
Communications Biology (2023)
-
High-speed atomic force microscopy: extracting high-resolution information through image analysis
Biophysical Reviews (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.