Abstract
Gastrointestinal health depends on the adaptive immune system tolerating the foreign proteins in food1,2. This tolerance is paradoxical because the immune system normally attacks foreign substances by generating inflammation. Here we addressed this conundrum by using a sensitive cell enrichment method to show that polyclonal CD4+ T cells responded to food peptides, including a natural one from gliadin, by proliferating weakly in secondary lymphoid organs of the gut–liver axis owing to the action of regulatory T cells. A few food-specific T cells then differentiated into T follicular helper cells that promoted a weak antibody response. Most cells in the expanded population, however, lacked canonical T helper lineage markers and fell into five subsets dominated by naive-like or T follicular helper-like anergic cells with limited capacity to form inflammatory T helper 1 cells. Eventually, many of the T helper lineage-negative cells became regulatory T cells themselves through an interleukin-2-dependent mechanism. Our results indicate that exposure to food antigens causes cognate CD4+ naive T cells to form a complex set of noncanonical hyporesponsive T helper cell subsets that lack the inflammatory functions needed to cause gut pathology and yet have the potential to produce regulatory T cells that may suppress it.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Single-cell RNA-sequencing data are available in the Gene Expression Omnibus (accession number GSE202423). Source data are provided with this paper.
References
Renz, H. et al. Food allergy. Nat. Rev. Dis. Primers 4, 17098 (2018).
Green, P. H., Lebwohl, B. & Greywoode, R. Celiac disease. J. Allergy Clin. Immunol. 135, 1099–1106 (2015).
Pabst, O. & Mowat, A. M. Oral tolerance to food protein. Mucosal Immunol. 5, 232–239 (2012).
Liu, E. G., Yin, X., Swaminathan, A. & Eisenbarth, S. C. Antigen-presenting cells in food tolerance and allergy. Front. Immunol. 11, 616020 (2020).
Mowat, A. M. To respond or not to respond - a personal perspective of intestinal tolerance. Nat. Rev. Immunol. 18, 405–415 (2018).
Battaglia, M., Gianfrani, C., Gregori, S. & Roncarolo, M. G. IL-10-producing T regulatory type 1 cells and oral tolerance. Ann. N. Y. Acad. Sci. 1029, 142–153 (2004).
Weiner, H. L., da Cunha, A. P., Quintana, F. & Wu, H. Oral tolerance. Immunol. Rev. 241, 241–259 (2011).
Curotto de Lafaille, M. A. et al. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity 29, 114–126 (2008).
Hadis, U. et al. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34, 237–246 (2011).
Kim, K. S. et al. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 351, 858–863 (2016).
Hataye, J., Moon, J. J., Khoruts, A., Reilly, C. & Jenkins, M. K. Naive and memory CD4+ T cell survival controlled by clonal abundance. Science 312, 114–116 (2006).
Marzo, A. L. et al. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat. Immunol. 6, 793–799 (2005).
Moon, J. J. et al. Naive CD4+ T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27, 203–213 (2007).
Nelson, R. W. et al. T cell receptor cross-reactivity between similar foreign and self peptides influences naive cell population size and autoimmunity. Immunity 42, 95–107 (2015).
Moser, B. CXCR5, the defining marker for follicular B helper T (TFH) cells. Front. Immunol. 6, 296 (2015).
Jiang, S. & Dong, C. A complex issue on CD4+ T-cell subsets. Immunol. Rev. 252, 5–11 (2013).
Kotov, J. A. & Jenkins, M. K. Cutting edge: T cell-dependent plasmablasts form in the absence of single differentiated CD4+ T cell subsets. J. Immunol. 202, 401–405 (2019).
Kalekar, L. A. et al. CD4+ T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat. Immunol. 17, 304–314 (2016).
Rees, W. et al. An inverse relationship between T cell receptor affinity and antigen dose during CD4+ T cell responses in vivo and in vitro. Proc. Natl Acad. Sci. USA 96, 9781–9786 (1999).
Pepper, M., Pagán, A. J., Igyártó, B. Z., Taylor, J. J. & Jenkins, M. K. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35, 583–595 (2011).
Grover, H. S. et al. The Toxoplasma gondii peptide AS15 elicits CD4 T cells that can control parasite burden. Infect. Immun. 80, 3279–3288 (2012).
Geginat, G., Schenk, S., Skoberne, M., Goebel, W. & Hof, H. A novel approach of direct ex vivo epitope mapping identifies dominant and subdominant CD4 and CD8 T cell epitopes from Listeria monocytogenes. J. Immunol. 166, 1877–1884 (2001).
Lycke, N. The mechanism of cholera toxin adjuvanticity. Res. Immunol. 148, 504–520 (1997).
Matloubian, M. et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360 (2004).
Ohtani, N. & Kawada, N. Role of the gut–liver axis in liver inflammation, fibrosis, and cancer: a special focus on the gut microbiota relationship. Hepatol. Commun. 3, 456–470 (2019).
Marshall, H. D. et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4+ cell properties during viral infection. Immunity 35, 633–646 (2011).
MacMicking, J. D. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25, 601–609 (2004).
Moran, A. E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Longhi, M. P. et al. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 206, 1589–1602 (2009).
Fontenot, J. D., Rasmussen, J. P., Gavin, M. A. & Rudensky, A. Y. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6, 1142–1151 (2005).
Coombes, J. L. et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta– and retinoic acid–dependent mechanism. J. Exp. Med. 204, 1757–1764 (2007).
Cong, Y., Weaver, C. T. & Elson, C. O. The mucosal adjuvanticity of cholera toxin involves enhancement of costimulatory activity by selective up-regulation of B7.2 expression. J. Immunol. 159, 5301–5308 (1997).
Bromander, A. K., Kjerrulf, M., Holmgren, J. & Lycke, N. Cholera toxin enhances alloantigen presentation by cultured intestinal epithelial cells. Scand. J. Immunol. 37, 452–458 (1993).
Wong, H. S. et al. A local regulatory T cell feedback circuit maintains immune homeostasis by pruning self-activated T cells. Cell 184, 3981–3997 (2021).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Liao, W., Lin, J. X., Wang, L., Li, P. & Leonard, W. J. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 12, 551–559 (2011).
Ballesteros-Tato, A. et al. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36, 847–856 (2012).
Johnston, R. J., Choi, Y. S., Diamond, J. A., Yang, J. A. & Crotty, S. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 209, 243–250 (2012).
Kiner, E. et al. Gut CD4+ T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat Immunol. 22, 216–228 (2021).
Sumida, T. S. et al. Type I interferon transcriptional network regulates expression of coinhibitory receptors in human T cells. Nat. Immunol. 23, 632–642 (2022).
Trefzer, A. et al. Dynamic adoption of anergy by antigen-exhausted CD4+ T cells. Cell Rep. 34, 108748 (2021).
Mohrs, K., Wakil, A. E., Killeen, N., Locksley, R. M. & Mohrs, M. A two-step process for cytokine production revealed by IL-4 dual-reporter mice. Immunity 23, 419–429 (2005).
McSorley, S. J., Asch, S., Costalonga, M., Reinhardt, R. L. & Jenkins, M. K. Tracking Salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity 16, 71–83 (2002).
Leehan, K. M. & Koelsch, K. A. T cell ELISPOT: for the identification of specific cytokine-secreting T cells. Methods Mol. Biol. 1312, 427–434 (2015).
Qiu, Z. & Sheridan, B. S. Isolating lymphocytes from the mouse small intestinal immune system. J. Vis. Exp. https://doi.org/10.3791/57281 (2018).
Ertelt, J. M. et al. Selective priming and expansion of antigen-specific Foxp3−CD4+ T cells during Listeria monocytogenes infection. J. Immunol. 182, 3032–3038 (2009).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Bonecchi, R. et al. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 187, 129–134 (1998).
Glatigny, S., Duhen, R., Oukka, M. & Bettelli, E. Cutting edge: loss of α4 integrin expression differentially affects the homing of Th1 and Th17 cells. J. Immunol. 187, 6176–6179 (2011).
Swarnalekha, N. et al. T resident helper cells promote humoral responses in the lung. Sci. Immunol. 6, eabb6808 (2021).
Acknowledgements
We acknowledge J. Walter and C. Elwood for help with maintaining mice and the University of Minnesota Flow Cytometry Resource for maintenance of flow cytometers used in these studies. This work was supported by the NIH grant P01 AI035296 and R01 AI27998.
Author information
Authors and Affiliations
Contributions
S.-W.H. and M.K.J. designed experiments. S.-W.H., P.D.K. and K.C.O. carried out experiments. T.D. produced tetramers. A.H. and D.L.M. carried out sequencing data analysis. S.-W.H. and M.K.J. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Kenya Honda, Oliver Pabst and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Gliadin antibody production after feeding depends on Tfh cells.
Gliadin antibody amounts were determined by Enzyme-Linked Immunosorbent Assay in serial dilutions of serum from Bcl6fl/fl (n = 6 mice) or LckCre Bcl6fl/fl mice (n = 6 mice) on a gliadin-containing diet, wild-type C57BL/6 mice on a gliadin-free diet (n = 2 mice), or C57BL/6 mice on a gliadin-free diet mice 2 weeks after subcutaneous injection of gliadin in complete Freund’s adjuvant (n = 2 mice). Wild-type and Bcl6fl/fl mice have the capacity to produce Bcl-6 and TFH cells. LckCre Bcl6fl/fl mice have a T cell-specific defect in Bcl-6 and cannot produce TFH cells. Values for each dilution from Bcl6fl/fl or LckCre Bcl6fl/fl mice were compared by Student’s t-test. The data show that TFH-sufficient mice on a gliadin-containing diet make small amounts of gliadin antibody, while TFH-deficient mice do not.
Extended Data Fig. 2 THlin− cells induced by peptide feeding are stable after cessation of peptide feeding and lack markers of TH2, Tr1, and TH3 cells.
a, Flow cytometry plots of 2W:I-Ab tetramer-binding CD4+ T cells from mice fed with 2W peptide 3 times and analyzed 2 days or 1 week after cessation of feeding, with a gate on Treg cells. b, Percentages of Treg (left) or THlin− (right) cells in 2W:I-Ab tetramer-binding populations from mice fed with 2W peptide 3 times and analyzed at the indicated times after cessation of feeding (n = 6 mice for 2 days, n = 5 mice for 1 wk, n = 4 mice for 2 wks, n = 3 mice for 3 wks). c, Flow cytometry plots of 2W:I-Ab tetramer-binding Thlin− cells from Il4CD2 (left plot) or wild-type (middle and right plots) mice fed with 2W peptide 3 times and analyzed 2 days after cessation of feeding for expression of human CD2 (a marker of IL-4 and TH2 cells in Il4CD2 mice), LAG-3 (a marker of Tr1 cells), or LAP (a marker of TH3 cells) (n = 4 or 5 mice). The scatterplot shows the percentages of cells with these markers in the 2W:I-Ab tetramer-binding Thlin− populations from individual mice
Extended Data Fig. 3 Experimental scheme used to identify CD4+ T cell subsets and a heatmap of protein expression assessed by flow cytometry.
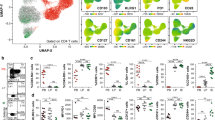
a, Experimental scheme used to identify 2W:I-Ab, TOXO:I-Ab, or LLO:I-Ab tetramer-binding THlin− cells from CD45.1 mice fed the peptides on days 0, 2, and 4 and analyzed 2 days after the last feeding, 2W:I-Ab, TOXO:I-Ab, or LLO:I-Ab tetramer-binding TH1 and TH17 cells from mice immunized 7 days earlier with the peptides in complete Freund’s adjuvant (CFA), or tetramer-negative Treg, naïve, TFH, and anergic T cells. b, Heatmap of the log2 values of mean fluorescence intensity (MFI) for each PE-labeled antibody for a given T cell subset divided by the MFI for that antibody for naïve T cells. These normalized values were used for the PCA shown in Fig. 2a.
Extended Data Fig. 4 Flow cytometry histograms of protein expression by CD4+ T cell subsets.
a, Histograms of antibody staining of the indicated proteins on or in the indicated subsets identified in Extended Data Fig. 3a. b, Histograms of antibody staining of the indicated proteins on or in Glp:I-Ab tetramer-binding THlin− cells identified as in Extended Data Fig. 3a from mice on a gliadin-containing diet for 1 week. The other subsets were identified as in Extended Data Fig. 3a except that the TH1/TH17 cells were isolated from the non-tetramer-binding CD4+ T cell population.
Extended Data Fig. 6 RNA sequencing data used for definition of clusters of CD4+ T cells from mice on a gliadin-containing diet.
a, Single-cell RNA sequencing analysis of Glp:I-Ab tetramer-binding T cells from mice exposed to a gliadin-containing diet for 1 week (Gliadin), 2W:I-Ab tetramer-binding T cells from mice infected with 2W-expressing Listeria monocytogenes (Lm), or naïve CD4+ T cells (Naïve). UMAPs identifying 9 T cell clusters in merged populations (left panel) and in each population (right panel) are shown. b, c, Expression levels of signature genes from each cluster are shown as violin plots (b) or a heat map (c). Note that the total Glp:I-Ab tetramer-binding T cell population was included in the analysis not just the CD44hi subset. In this analysis, clusters 1 and 2 contained naïve T cells defined by expression of Sell, Ccr7, and Ly6C1. The Lm-induced 2W:I-Ab-specific effector cell population, known to contain TH1 and TFH cells20, consisted of cells in clusters 0, 3, 5, and 6. Clusters 0 and 3 contained TH1 cells as evidenced by expression of the TH1 markers Cxcr3 and Itga4, with cells in cluster 3 expressing markers of more advanced TH1 differentiation (Ccr2, Nkg7, and Ly6C2)49,50,51. Cluster 5 contained TFH cells as evidenced by expression of the markers Bcl6 and Cxcr5. Cluster 6 was defined by cells expressing interferon-induced genes (Gbp2, Stat1, Ifi47, Ly6A, and Igtp)27 and may contain TFH-related cells based on proximity in UMAP space to cluster 5. The Glp:I-Ab tetramer-binding population from mice on a gliadin-containing diet contained some naïve cells in clusters 1 and 2 and cells in clusters 4, 7, and 8 that were not found in the Lm-induced effector population. Cluster 7 contained Treg cells based on expression of Foxp3, while the cells in cluster 8 were TFH-like cells based on expression of Bcl6 and Cxcr5 but were different from the genuine cluster 5 TFH cells induced by Listeria infection. The cells in cluster 4 were defined by expression of naïve cell markers Sell and Ccr7 and markers of TCR signaling such as Nr4a1, which were also expressed by the Treg cells in cluster 7 and the TFH-like cells in cluster 8. Glp:I-Ab tetramer-binding cells in clusters 4 and 8 also expressed Izumor1. Cluster 4 in the Glp:I-Ab tetramer-binding population shown in Extended Data Fig. 6a likely corresponds to THlin− naïve-like cluster 0 or 1 in the peptide-feeding experiment shown in Fig. 2b, while cluster 8 in the Glp:I-Ab tetramer-binding population likely corresponds to TFH-like cluster 3 in the peptide feeding experiment. The minor cluster 8 (THlin− type 1 interferon-signaled) and cluster 9 (THlin− TFH-like TCR signaling) subsets that were detected in the peptide feeding experiment (Fig. 2b) were not detected in the Glp:I-Ab tetramer-binding population shown in Extended Data Fig. 6a, perhaps because fewer cells were analyzed in this case. Thus, the cognate T cell population induced by an antigen in a natural diet and that induced by peptide feeding by gavage contained Treg cells and THlin− cells with naïve-like and TFH-like subsets.
Supplementary information
Rights and permissions
About this article
Cite this article
Hong, SW., Krueger, P.D., Osum, K.C. et al. Immune tolerance of food is mediated by layers of CD4+ T cell dysfunction. Nature 607, 762–768 (2022). https://doi.org/10.1038/s41586-022-04916-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-022-04916-6
This article is cited by
-
Therapeutic induction of antigen-specific immune tolerance
Nature Reviews Immunology (2024)
-
Cleaving an epithelial path to food tolerance
Cell Research (2023)
-
Regulatory T cells in the face of the intestinal microbiota
Nature Reviews Immunology (2023)
-
The Functional Adaptability of Hyporesponsive T Cells and Its Impact on Transplant Outcomes
Current Transplantation Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.