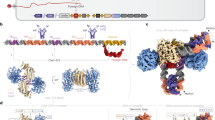
Abstract
Prokaryotes adapt to challenges from mobile genetic elements by integrating spacers derived from foreign DNA in the CRISPR array1. Spacer insertion is carried out by the Cas1–Cas2 integrase complex2,3,4. A substantial fraction of CRISPR–Cas systems use a Fe–S cluster containing Cas4 nuclease to ensure that spacers are acquired from DNA flanked by a protospacer adjacent motif (PAM)5,6 and inserted into the CRISPR array unidirectionally, so that the transcribed CRISPR RNA can guide target searching in a PAM-dependent manner. Here we provide a high-resolution mechanistic explanation for the Cas4-assisted PAM selection, spacer biogenesis and directional integration by type I-G CRISPR in Geobacter sulfurreducens, in which Cas4 is naturally fused with Cas1, forming Cas4/Cas1. During biogenesis, only DNA duplexes possessing a PAM-embedded 3′-overhang trigger Cas4/Cas1–Cas2 assembly. During this process, the PAM overhang is specifically recognized and sequestered, but is not cleaved by Cas4. This ‘molecular constipation’ prevents the PAM-side prespacer from participating in integration. Lacking such sequestration, the non-PAM overhang is trimmed by host nucleases and integrated to the leader-side CRISPR repeat. Half-integration subsequently triggers PAM cleavage and Cas4 dissociation, allowing spacer-side integration. Overall, the intricate molecular interaction between Cas4 and Cas1–Cas2 selects PAM-containing prespacers for integration and couples the timing of PAM processing with the stepwise integration to establish directionality.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM density maps that support the findings of this study have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-23839 (PAM/PAM prespacer bound), EMD-23840 (PAM/non-PAM prespacer bound), EMD-23843 (full-integration complex), EMD-23845 (half-integration complex, Cas4 still blocking the PAM side), EMD-23849 (half-integration complex, Cas4 dissociated) and EMD-23847 (sub-complex). The coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers 7MI4 (PAM/PAM prespacer-bound), 7MI5 (PAM/non-PAM prespacer-bound), 7MI9 (full integration), 7MIB (half integration, Cas4 still blocking the PAM side), 7MID (sub-complex). MiSeq sequencing data that support analysis of in vivo prespacer integration have been deposited in the European Nucleotide Archive (ENA) under accession number PRJEB41616. Plasmids used in this study are available upon request.
References
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Yosef, I., Goren, M. G. & Qimron, U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40, 5569–5576 (2012).
Nuñez, J. K. et al. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat. Struct. Mol. Biol. 21, 528–534 (2014).
Nuñez, J. K., Lee, A. S., Engelman, A. & Doudna, J. A. Integrase-mediated spacer acquisition during CRISPR–Cas adaptive immunity. Nature 519, 193–198 (2015).
Mojica, F. J., Diez-Villasenor, C., Garcia-Martinez, J. & Almendros, C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155, 733–740 (2009).
Marraffini, L. A. & Sontheimer, E. J. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463, 568–571 (2010).
Vink, J. N. A. et al. Direct visualization of native CRISPR target search in live bacteria reveals cascade DNA surveillance mechanism. Mol. Cell 77, 39–50.e10 (2020).
Nuñez, J. K., Harrington, L. B., Kranzusch, P. J., Engelman, A. N. & Doudna, J. A. Foreign DNA capture during CRISPR–Cas adaptive immunity. Nature 527, 535–538 (2015).
Wright, A. V. & Doudna, J. A. Protecting genome integrity during CRISPR immune adaptation. Nat. Struct. Mol. Biol. 23, 876–883 (2016).
Wright, A. V. et al. Structures of the CRISPR genome integration complex. Science 357, 1113–1118 (2017).
Budhathoki, J. B. et al. Real-time observation of CRISPR spacer acquisition by Cas1–Cas2 integrase. Nat. Struct. Mol. Biol. 27, 489–499 (2020).
Xiao, Y., Ng, S., Nam, K. H. & Ke, A. How type II CRISPR-Cas establish immunity through Cas1–Cas2-mediated spacer integration. Nature 550, 137–141 (2017).
Kim, S. et al. Selective loading and processing of prespacers for precise CRISPR adaptation. Nature 579, 141–145 (2020).
Li, M., Wang, R., Zhao, D. & Xiang, H. Adaptation of the Haloarcula hispanica CRISPR–Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 42, 2483–2492 (2014).
Liu, T. et al. Coupling transcriptional activation of CRISPR-Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 45, 8978–8992 (2017).
Shiimori, M., Garrett, S. C., Graveley, B. R. & Terns, M. P. Cas4 nucleases define the PAM, length, and orientation of DNA fragments integrated at CRISPR loci. Mol. Cell 70, 814–824.e6 (2018).
Kieper, S. N. et al. Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep. 22, 3377–3384 (2018).
Almendros, C., Nobrega, F. L., McKenzie, R. E. & Brouns, S. J. J. Cas4–Cas1 fusions drive efficient PAM selection and control CRISPR adaptation. Nucleic Acids Res. 47, 5223–5230 (2019).
Lemak, S. et al. Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus. J. Am. Chem. Soc. 135, 17476–17487 (2013).
Lemak, S. et al. The CRISPR-associated Cas4 protein Pcal_0546 from Pyrobaculum calidifontis contains a [2Fe-2S] cluster: crystal structure and nuclease activity. Nucleic Acids Res. 42, 11144–11155 (2014).
Zhang, J., Kasciukovic, T. & White, M. F. The CRISPR associated protein Cas4 is a 5′ to 3′ DNA exonuclease with an iron–sulfur cluster. PLoS ONE 7, 0047232 (2012).
Lee, H., Dhingra, Y. & Sashital, D. G. The Cas4–Cas1–Cas2 complex mediates precise prespacer processing during CRISPR adaptation. eLife 8, e44248 (2019).
Lee, H., Zhou, Y., Taylor, D. W. & Sashital, D. G. Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol. Cell 70, 48–59.e5 (2018).
Xiao, Y., Luo, M., Dolan, A. E., Liao, M. & Ke, A. Structure basis for RNA-guided DNA degradation by Cascade and Cas3. Science 361, aat0839 (2018).
Shah, S. A., Erdmann, S., Mojica, F. J. & Garrett, R. A. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 10, 891–899 (2013).
Jia, N. et al. Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection. Proc. Natl Acad. Sci. USA 116, 24507–24516 (2019).
Nuñez, J. K., Bai, L., Harrington, L. B., Hinder, T. L. & Doudna, J. A. CRISPR immunological memory requires a host factor for specificity. Mol. Cell 62, 824–833 (2016).
Ramachandran, A., Summerville, L., Learn, B. A., DeBell, L. & Bailey, S. Processing and integration of functionally oriented prespacers in the Escherichia coli CRISPR system depends on bacterial host exonucleases. J. Biol. Chem. 295, 3403–3414 (2020).
Levy, A. et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 520, 505–510 (2015).
Modell, J. W., Jiang, W. & Marraffini, L. A. CRISPR–Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544, 101–104 (2017).
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
Hudaiberdiev, S. et al. Phylogenomics of Cas4 family nucleases. BMC Evol. Biol. 17, 232 (2017).
Pourcel, C. et al. CRISPRCasdb a successor of CRISPRdb containing CRISPR arrays and cas genes from complete genome sequences, and tools to download and query lists of repeats and spacers. Nucleic Acids Res. 48, D535–D544 (2020).
Pruitt, K. D., Tatusova, T. & Maglott, D. R. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33, D501–D504 (2005).
Benson, D. A. et al. GenBank. Nucleic Acids Res. 46, D41–D47 (2018).
Sayers, E. W. et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37, D5–D15 (2009).
Arndt, D. et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21 (2016).
Mitchell, A. L. et al. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 48, D570–D578 (2020).
Chen, I. A. et al. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res. 45, D507–D516 (2017).
Paez-Espino, D. et al. IMG/VR v.2.0: an integrated data management and analysis system for cultivated and environmental viral genomes. Nucleic Acids Res. 47, D678–D686 (2019).
Soto-Perez, P. et al. CRISPR–Cas system of a prevalent human gut bacterium reveals hyper-targeting against phages in a human virome catalog. Cell Host Microbe 26, 325–335.e325 (2019).
Group, N. H. W. et al. The NIH Human Microbiome Project. Genome Res. 19, 2317–2323 (2009).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e620 (2019).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Deveau, H. et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 (2008).
Almendros, C., Guzman, N. M., Diez-Villasenor, C., Garcia-Martinez, J. & Mojica, F. J. Target motifs affecting natural immunity by a constitutive CRISPR–Cas system in Escherichia coli. PLoS ONE 7, e50797 (2012).
Lange, S. J., Alkhnbashi, O. S., Rose, D., Will, S. & Backofen, R. CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res. 41, 8034–8044 (2013).
Alkhnbashi, O. S. et al. CRISPRstrand: predicting repeat orientations to determine the crRNA-encoding strand at CRISPR loci. Bioinformatics 30, i489–i496 (2014).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
McKenzie, R. E., Almendros, C., Vink, J. N. A. & Brouns, S. J. J. Using CAPTURE to detect spacer acquisition in native CRISPR arrays. Nat. Protoc. 14, 976–990 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Xu, K., Zang, X., Peng, M., Zhao, Q. & Lin, B. Magnesium lithospermate B downregulates the levels of blood pressure, inflammation, and oxidative stress in pregnant rats with hypertension. Int. J. Hypertens. 2020, 6250425 (2020).
Acknowledgements
This work is supported by the Netherlands Organization for Scientific Research (NWO) VICI grant (VI.C.182.027) to S.J.J.B. and the National Institutes of Health (NIH) grant (GM118174) to A.K. This work made use of the Cornell Center for Materials Research Shared Facilities which are supported through the NSF MRSEC program (DMR-1719875). We thank S. N. Kieper, R. Miojevic, M. Ramos, G. Schuler and K. Spoth for helpful discussions, advice and technical assistance.
Author information
Authors and Affiliations
Contributions
A.K., S.J.J.B., C.H. and C.A. designed the research. C.H. is responsible for biochemistry and cryo-EM reconstructions; C.A., J.N.A.V., A.R.C. and A.C.H. are responsible for in vivo and bioinformatics analyses; K.H.N. and C.H. are responsible for structure building and refinement; and S.R.B. assisted with cryo-EM work. A.K. and C.H. wrote the manuscript with input from S.J.J.B., J.N.A.V. and A.R.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Martin Jinek, Lennart Randau and Malcolm White for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Reconstitution and characterization of the GsuCas4/Cas1-Cas2 complex.
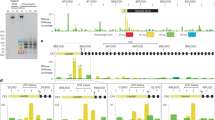
a. Active site substitution in Cas4 nuclease center (H48G, D100A) reduced in vivo spacer acquisition efficiency dramatically. Left three panels display the WebLogo of PAM code from spacers integrated by each Cas4/1-2 variant. Rightmost panel displays the number of deep-sequencing reads that confirm spacer integration. b–d. GsuCas4/1 purification analyzed by SDS-PAGE, coloring from the Fe-S cluster, and SEC profile, respectively. e,f. Affinity purification of GsuCas2, SDS-PAGE, and SEC analysis, respectively. g. GST pull-down experiments revealing the physical interaction between GsuCas4/1 and GsuCas2, with or without prespacer present. h. Metal ion dependency in PAM cleavage reaction. i. Biochemistry showing Cas4/1-2 specifically cleaves the PAM-embedded 3′-overhang in prespacer. j. PAM-cleavage specificity is lost over time, presumably due to Fe-S oxidation in Cas4. k. SEC profile of GsuCas4/Cas1-Cas2, alone or programmed with different prespacer substrates. PAM-containing prespacers drive high-order complex formation. l. Cryo-electron micrographs of three different complexes, with corresponding preliminary 2D averages to investigate sample quality.
Extended Data Fig. 2 In-depth analysis of the dual-PAM prespacer bound GsuCas4/Cas1-Cas2 structure.
a. Comparison between the current 3.2 Å cryo-EM reconstruction with the previous negative staining reconstruction of the B. hal Cas4/1-2 complex (EMDB 20131)22. b–d. Pairwise alignment between GsuCas4/Cas1-Cas2/prespacer and EcoCas1-Cas2/prespacer8,31 (PDB 5DS4), EfaCas1-Cas2/prespacer12 (PDB 5XVN), and EfaCas1-Cas2/full-integration12 (PDB 5XVO), respectively. Alignments details are noted on the figure panel. Inset: the C-terminal tail of Cas2 plays similar roles in G. sul and E. fae structures in mediating edge-stacking with both Cas2 and Cas1. e. PAM was processed similarly in 22-bp or 26-bp mid-duplex containing prespacer by GsuCas4/Cas1-Cas2. f. SEC profile was similar when the two different prespacers were used to assemble the complex. g. Validation that prespacers containing a 22-bp mid-duplex are actively acquired in vivo. N=3 biologically independent assays were evaluated by PCR detection as shown, as well as relative percentages of expanded and non-expanded bands. Data presented as mean ± s.e.m.
Extended Data Fig. 3 Flow-chart of the cryo-EM single particle reconstruction of the dual-PAM prespacer bound GsuCas4/Cas1-Cas2.
a. Cryo-EM reconstruction workflow for the dual-PAM prespacer bound Cas4/1-2 complex. b. Cryo-EM density of the dual-PAM prespacer bound Cas4/1-2 complex, colored according to local resolution (top). The viewing direction distribution plot (middle) and FSC curves (bottom) for data processing. c. Representative EM densities for Cas2, Cas4, and Cas1, superimposed with their corresponding structural model.
Extended Data Fig. 4 In-depth GsuCas4/Cas1-Cas2 interface analysis and structure-guided mutagenesis attempt to switch PAM specificity.
a. Overall dual-PAM structure. Insets: zoom-ins of interface between Cas4 and the two neighboring Cas1s. Cas4 connects to the non-catalytic Cas1 through a 20-amino acid fusion linker (colored in yellow), which mediates the dynamic docking and dissociation of Cas4. b. Surface electrostatic potential. Left inset: Cas2 contacts to the mid-duplex; Right inset: Cas1 end-stacking to the mid-duplex. Residues responsible for guiding the 3’-overhang are also shown. Cas1-Cas2 was found to specify a 22-bp mid-duplex rather than a 26-bp mid-duplex as defined by the integration assay; an additional two base-pairs are unwound from each end, and the mid-duplex is end-stacked by the N-terminal domain of the catalytic Cas1s on opposite ends. The 22-bp specification and the limited end-unwinding activity was previously observed in EfaCas1-Cas211,12. c. Cas1-Cas2 and Cas4-Cas2 interfaces. Top inset: the highly conserved C-terminus of Cas2 inserting into a hydrophobic pocket in Cas1, stabilizing complex formation. Bottom inset: the ceiling helix of Cas4 (aa 39–50) makes extensive polar contacts with a helix in Cas2 (aa 42–53). d. SEC, SDS-PAGE, and urea-PAGE analyses of the prespacer-bound complex used in cryo-EM analysis. They reveal the molecular weight, protein integrity, and prespacer integrity, respectively. For example, urea-PAGE reveals the PAM-overhang is not cleaved inside the Cas4/1-2 complex. e. Modeling the impact on PAM recognition by introducing the equivalent residues of E18 and S191 in P. fur Cas4 into G. sul Cas4 (E18Y and S191A substitutions). Specific atom changes in A-to-G switching (N6O substitution and N2 amine addition) are highlighted in colored balls. The steric clashes (lightening arrows) to PfuPAM (3’-GGN in the 3’-overhang) are expected to be partially relieved when substitutions are in place. f. Impact of E18Y and S191A substitutions on PAM cleavage activity. g. In vivo spacer acquisition assay results for the wild type and PAM-specificity Cas4 mutants. While E18Y/S191A Cas4 showed compromised Gsu-PAM (TTN) prespacer integration, it was able to support integration of Pfu-PAM (CCN) containing prespacers in vivo. N = 3 biological independent assays were analyzed by PCR and the band quantification revealed integration efficiency. Data presented as mean ± s.e.m.
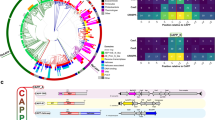
Extended Data Fig. 5 In-depth analysis of the structure and sequence conservation in Cas4.
a. Superposition of GsuCas4 with a standalone Cas419,20, and the nuclease domains in helicase-nuclease fusion proteins AddAB32, AdnAB26, RecBCD33, and eukaryotic Dna234. The caging of the ssDNA substrate and the arrangement of the Fe-S cluster and the catalytic triad are conserved themes. Interestingly, the Cas4 structure aligns poorly with the RecB nuclease in RecBCD; it agrees better with the RecB-like fold in RecC instead. b, c. Sequence alignment of GsuCas4, GsuCas1, and PfuCas4 with their close homologs. Based on the structural analysis, we marked the residues important for subunit interaction, substrate binding, catalysis and Fe-S cluster formation. d. Quality of the purified GsuCas4 mutants that carry the PAM-recognition residues from PfuCas4. These mutants were used in the structure-guided PAM-switching experiments in Extended Data Fig. 4.
Extended Data Fig. 6 Cryo-EM single particle reconstruction of the single-PAM prespacer bound GsuCas4/Cas1-Cas2.
a. Flow-chart of the cryo-EM single particle reconstruction process that led to the reconstruction of two major snapshots. Left: Asymmetrical PAM/Non-PAM prespacer bound Cas4/1-2 complex. Right: That of the sub complex lacking (Cas4/1)2 on the non-PAM side. b. Cryo-EM density of the two reconstructions colored according to local resolution (top); viewing direction distribution plot (middle); and FSC curves (bottom). c. Superposition of the PAM side and non-PAM side densities showing that Cas4 density is largely missing at the non-PAM side, and the non-PAM 3’-overhang is largely disordered. Only the first four nucleotides of the non-PAM 3′-overhang can be traced in the density, along a similar path as in the PAM-side.
Extended Data Fig. 7 In vitro assays to distinguish integration directionality.
a, b. Biochemistry showing that GsuCas4/1-2 is unable to integrate prespacer into the linear form of leader-repeat DNA. c. Successful prespacer integration into a leader-repeat containing plasmid by Cas4/1-2. d. The leader-repeat sequence cloned into the plasmid. We cleaved the leader-repeat sequence via the EcoRI and XhoI sites after the integration assay to further resolve the integration directionality on urea-PAGE. e. Schematic diagram explaining how the integration directionality can be resolved based on the fluorescent ssDNA sizes. f. Integration profile in urea-PAGE when both overhangs are integration-ready (7-nt long). Results showed that from the leader-repeat point of view, integration preferentially initiates from the leader-side, as the spacer-side integration trails after the leader-side integration in the time-course experiment. From the prespacer point of view, the integration directionality is scrambled. Each integration band contains two overlapping fluorescent signals. g. Native PAGE showing that in the concentration-gradient experiment, complex formation between Cas4/1-2 and prespacer takes place in a stepwise and PAM-dependent fashion.
Extended Data Fig. 8 In-depth analysis of the mechanistic coupling between half-integration and PAM cleavage by Cas4.
a. Time-course experiment showing ExoI trims PAM and non-PAM overhangs differently. The non-PAM 3′-overhang was trimmed to within one nucleotide of the preferred length, 7 nt. The PAM-side 3′-overhang was protected by the footprint of Cas4 in the same reaction. b. Time-course experiment resolving the order of events from prespacer processing to full integration. Using the Cas4/1-2 (left set) and Cas4/1-2 plus ExoI (middle set) lanes as controls, the right set of experiment shows ExoI trimming triggers the integration of the non-PAM overhang into the leader-proximal target DNA. This is followed by the stimulation of PAM cleavage, and then the full integration from PAM-overhang to spacer-side target. c. Temperature-dependency of PAM cleavage and spacer-side integration. d. Side-by-side comparison of PAM cleavage at 50 °C, prespacer alone or programmed to the half-integrated state. e. Quantification of the cleaved band in c. and d. revealing the elevated PAM cleavage and full integration when leader-side integration already took place. Data were collected from N = 3 biologically independent experiments and presented with mean ± s.e.m. Statistical significance was assessed by two-tailed t-test, with the exact P values displayed. f. Salt-dependency of PAM cleavage and full integration. g–i. Optimization of full integration reaction by defining its time course, Cas2-dependency, and pH-dependency, respectively. j. Defining pH-dependency of PAM cleavage by Cas4. k. SEC analysis of the Cas4/1-2 complex programmed with the half-integration product mimic. Samples in the integrated complex peak was used for cryo-EM data collection and single particle reconstruction. l, Schematics of the half-integration product mimic annealed from oligonucleotides. m. Urea-PAGE analysis of the SEC peak in k. revealing that Cas4/1-2 further catalyzed the full-integration reaction after binding to the half-integration mimic.
Extended Data Fig. 9 Cryo-EM single particle reconstruction of GsuCas4/Cas1-Cas2 programmed with a half-integration mimic.
a. Workflow of cryo-EM data processing. b. Overall cryo-EM density showing resolution distribution, viewing direction distribution plot, and FSC curves of three different snapshots. Left: half-integration, Cas4 disappeared; Middle: full-integration; Right: half-integration, Cas4 still blocking PAM-side.
Extended Data Fig. 10 In-depth analysis of the three snapshots captured from GsuCas4/Cas1-Cas2 programmed with a half-integration mimic.
a. Superposition of cryo-EM reconstructions to reveal the structural differences among three functional states. b. Orientation view of the full integration snapshot for additional interface analysis. The entire leader- repeat DNA is contacted in a quasi-symmetric fashion at the following four regions. c. Contacts from the two Cas1 subunits to the spacer-repeat DNA. The spacer-side DNA density is degenerate and DNA bending is not significant. The leader-recognition α-helix in the catalytic Cas1 is not inserted into the minor groove of the spacer-side DNA. d. The backbone of the central dyad of CRISPR repeat is contacted by the positive charges and a proline-rich motif on the ridge of the Cas2 dimer. e. Immediately adjacent to the catalytic loop, the linker connecting Cas4 to Cas1 is involved in DNA contact. A conserved PRPI motif is exposed upon Cas4 dissociation and is involved in DNA minor groove contact. f. The 4-bp leader region immediately upstream of the CRISPR repeat is favorably recognized and significantly bent upwards by the DNA minor groove insertion of a glycine-rich α-helix in Cas1. As previously revealed, this recognition leads to strong leader-proximal preference at the first half-integration reaction10,11,12. A pair of inverted repeats is found at the border region of the CRISPR repeat. This inverted repeat is recognized at the major groove region by the catalytic Histidine-containing loop in Cas112. g. Overall structure of the “Half-integration, Cas4 still blocking PAM-side” snapshot. This represents an early state, when Cas4 is still engaged in PAM recognition and the spacer-side leader-repeat is not allowed to enter into the integration site. h. The low-resolution EM density defines that the leader-repeat DNA preferentially contact a positively charged patch in Cas1. It should be noted that we are not able to define which specific DNA contact activates Cas4. This will require even higher temporal and spatial resolutions to resolve.
Supplementary information
Supplementary Figure 1
This file contains the uncropped gels shown in Figs 1, 3, 4 and Extended Data Figs 1, 2, 4, 7, 8.
Supplementary Tables
This file contains Supplementary Tables 1, 2, which contain lists of plasmids, primers and reagents used in the study.
Video 1
| Mechanism for Cas4-assisted prespacer biogenesis. This video illustrates the mechanism of Cas4-assisted prespacer biogenesis process. The animation made use of Pymol to interpolate structural transitions from one functional state to the next.
Video 2
| Mechanism for Cas4-assisted directional spacer integration.
Rights and permissions
About this article
Cite this article
Hu, C., Almendros, C., Nam, K.H. et al. Mechanism for Cas4-assisted directional spacer acquisition in CRISPR–Cas. Nature 598, 515–520 (2021). https://doi.org/10.1038/s41586-021-03951-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03951-z
This article is cited by
-
Insights into the inhibition of protospacer integration via direct interaction between Cas2 and AcrVA5
Nature Communications (2024)
-
Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays
Nature Structural & Molecular Biology (2023)
-
Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family
Communications Biology (2023)
-
Genome expansion by a CRISPR trimmer-integrase
Nature (2023)
-
Histones direct site-specific CRISPR spacer acquisition in model archaeon
Nature Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.