Abstract
Fibroblasts are polymorphic cells with pleiotropic roles in organ morphogenesis, tissue homeostasis and immune responses. In fibrotic diseases, fibroblasts synthesize abundant amounts of extracellular matrix, which induces scarring and organ failure. By contrast, a hallmark feature of fibroblasts in arthritis is degradation of the extracellular matrix because of the release of metalloproteinases and degrading enzymes, and subsequent tissue destruction. The mechanisms that drive these functionally opposing pro-fibrotic and pro-inflammatory phenotypes of fibroblasts remain unknown. Here we identify the transcription factor PU.1 as an essential regulator of the pro-fibrotic gene expression program. The interplay between transcriptional and post-transcriptional mechanisms that normally control the expression of PU.1 expression is perturbed in various fibrotic diseases, resulting in the upregulation of PU.1, induction of fibrosis-associated gene sets and a phenotypic switch in extracellular matrix-producing pro-fibrotic fibroblasts. By contrast, pharmacological and genetic inactivation of PU.1 disrupts the fibrotic network and enables reprogramming of fibrotic fibroblasts into resting fibroblasts, leading to regression of fibrosis in several organs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA-seq data used in this study have been deposited in the Gene Expression Omnibus (GEO) under the accession number GSE122334. The sequencing data from the ChIP–seq experiments have been submitted to the National Center for Biotechnology Information (NCBI) database under BioProject PRJNA480591; the project includes the following biosamples: SUB4300598, SUB4300595, SUB4300592, SUB4300591, SUB4300589, SUB4300587, SUB4300586, SUB4300583 and SUB4300579; the FASTQ data was uploaded to the NCBI Sequence Read Archive (SRA) under accession number SRP168244.
Change history
27 March 2019
The Source Data files for the figures were originally published in the wrong order, this has now been corrected.
References
Driskell, R. R. et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281 (2013).
Kalluri, R. & Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 6, 392–401 (2006).
Flavell, S. J. et al. Fibroblasts as novel therapeutic targets in chronic inflammation. Br. J. Pharmacol. 153, S241–S246 (2008).
Palumbo-Zerr, K. et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat. Med. 21, 150–158 (2015).
Ramming, A., Dees, C. & Distler, J. H. From pathogenesis to therapy — perspective on treatment strategies in fibrotic diseases. Pharmacol. Res. 100, 93–100 (2015).
Chakraborty, D. et al. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat. Commun. 8, 1130 (2017).
McInnes, I. B. & Schett, G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat. Rev. Immunol. 7, 429–442 (2007).
Parsonage, G. et al. Global gene expression profiles in fibroblasts from synovial, skin and lymphoid tissue reveals distinct cytokine and chemokine expression patterns. Thromb. Haemost. 90, 688–697 (2003).
Kiener, H. P. et al. Synovial fibroblasts self-direct multicellular lining architecture and synthetic function in three-dimensional organ culture. Arthritis Rheumatol. 62, 742–752 (2010).
Wynn, T. A. & Ramalingam, T. R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040 (2012).
Frank-Bertoncelj, M. et al. Epigenetically-driven anatomical diversity of synovial fibroblasts guides joint-specific fibroblast functions. Nat. Commun. 8, 14852 (2017).
Assassi, S. et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol. 67, 3016–3026 (2015).
Jolma, A. et al. DNA-binding specificities of human transcription factors. Cell 152, 327–339 (2013).
Scott, E. W., Simon, M. C., Anastasi, J. & Singh, H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265, 1573–1577 (1994).
McKercher, S. R. et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 15, 5647–5658 (1996).
Scanlan, M. J. et al. Molecular cloning of fibroblast activation protein α, a member of the serine protease family selectively expressed in stromal fibroblasts of epithelial cancers. Proc. Natl Acad. Sci. USA 91, 5657–5661 (1994).
Wu, M. et al. Identification of cadherin 11 as a mediator of dermal fibrosis and possible role in systemic sclerosis. Arthritis Rheumatol. 66, 1010–1021 (2014).
Uccelli, A., Moretta, L. & Pistoia, V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 8, 726–736 (2008).
Sheikh, H., Yarwood, H., Ashworth, A. & Isacke, C. M. Endo180, an endocytic recycling glycoprotein related to the macrophage mannose receptor is expressed on fibroblasts, endothelial cells and macrophages and functions as a lectin receptor. J. Cell Sci. 113, 1021–1032 (2000).
Soare, A., Ramming, A., Avouac, J. & Distler, J. H. W. Updates on animal models of systemic sclerosis. J. Scleroderma Relat. Disord. 1, 266–267 (2016).
Gabrielli, A., Avvedimento, E. V. & Krieg, T. Scleroderma. N. Engl. J. Med. 360, 1989–2003 (2009).
Sonnylal, S. et al. Postnatal induction of transforming growth factor β signaling in fibroblasts of mice recapitulates clinical, histologic, and biochemical features of scleroderma. Arthritis Rheumatol. 56, 334–344 (2007).
Györfi, A. H., Matei, A. E. & Distler, J. H. W. Targeting TGF-β signaling for the treatment of fibrosis. Matrix Biol. 68–69, 8–27 (2018).
Zeybel, M. et al. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat. Med. 18, 1369–1377 (2012).
Ramming, A., Druzd, D., Leipe, J., Schulze-Koops, H. & Skapenko, A. Maturation-related histone modifications in the PU.1 promoter regulate Th9-cell development. Blood 119, 4665–4674 (2012).
Van De Wiel, M. A. et al. Bayesian analysis of RNA sequencing data by estimating multiple shrinkage priors. Biostatistics 14, 113–128 (2013).
Bechtel, W. et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat. Med. 16, 544–550 (2010).
Noda, S. et al. Simultaneous downregulation of KLF5 and Fli1 is a key feature underlying systemic sclerosis. Nat. Commun. 5, 5797 (2014).
Cao, R. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043 (2002).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Christmann, R. B. et al. miR-155 in the progression of lung fibrosis in systemic sclerosis. Arthritis Res. Ther. 18, 155 (2016).
Migita, K. et al. TNF-α-induced miR-155 regulates IL-6 signaling in rheumatoid synovial fibroblasts. BMC Res. Notes 10, 403 (2017).
Vigorito, E. et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity 27, 847–859 (2007).
Pham, T. H. et al. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 41, 6391–6402 (2013).
Stephens, D. C. et al. Pharmacologic efficacy of PU.1 inhibition by heterocyclic dications: a mechanistic analysis. Nucleic Acids Res. 44, 4005–4013 (2016).
Munde, M. et al. The unusual monomer recognition of guanine-containing mixed sequence DNA by a dithiophene heterocyclic diamidine. Biochemistry 53, 1218–1227 (2014).
Farina, G., Lafyatis, D., Lemaire, R. & Lafyatis, R. A four-gene biomarker predicts skin disease in patients with diffuse cutaneous systemic sclerosis. Arthritis Rheumatol. 62, 580–588 (2010).
Sanchez-Lopez, E. et al. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor–insulin receptor substrate and STAT3 signaling. Oncogene 35, 2634–2644 (2016).
Scholten, D. et al. Migration of fibrocytes in fibrogenic liver injury. Am. J. Pathol. 179, 189–198 (2011).
Vollmann, E. H. et al. Identification of novel fibrosis modifiers by in vivo siRNA silencing. Mol. Ther. Nucleic Acids 7, 314–323 (2017).
Henderson, N. C. et al. Targeting of αV integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nat. Med. 19, 1617–1624 (2013).
Sakai, N. et al. LPA1-induced cytoskeleton reorganization drives fibrosis through CTGF-dependent fibroblast proliferation. FASEB J. 27, 1830–1846 (2013).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Gosselin, D. et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell 159, 1327–1340 (2014).
Bakri, Y. et al. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood 105, 2707–2716 (2005).
Chang, H. C. et al. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat. Immunol. 11, 527–534 (2010).
Anderson, K. L. et al. Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J. Immunol. 164, 1855–1861 (2000).
Rosenbauer, F. & Tenen, D. G. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7, 105–117 (2007).
Dweep, H., Sticht, C., Pandey, P. & Gretz, N. miRWalk – database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 44, 839–847 (2011).
Dweep, H. & Gretz, N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat. Methods 12, 697 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
van den Hoogen, F. et al. 2013 classification criteria for systemic sclerosis: an American college of rheumatology/European league against rheumatism collaborative initiative. Ann. Rheum. Dis. 72, 1747–1755 (2013).
Singal, A. K., Bataller, R., Ahn, J., Kamath, P. S. & Shah, V. H. ACG clinical guideline: alcoholic liver disease. Am. J. Gastroenterol. 113, 175–194 (2018).
EASL Clinical Practice Guidelines. Autoimmune hepatitis. J. Hepatol. 63, 971–1004 (2015).
Aletaha, D. et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann. Rheum. Dis. 69, 1580–1588 (2010).
Zheng, B., Zhang, Z., Black, C. M., de Crombrugghe, B. & Denton, C. P. Ligand-dependent genetic recombination in fibroblasts: a potentially powerful technique for investigating gene function in fibrosis. Am. J. Pathol. 160, 1609–1617 (2002).
Armaka, M. et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med. 205, 331–337 (2008).
Nutt, S. L., Metcalf, D., D’Amico, A., Polli, M. & Wu, L. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J. Exp. Med. 201, 221–231 (2005).
Rossi, A., Appelt-Menzel, A., Kurdyn, S., Walles, H. & Groeber, F. Generation of a three-dimensional full thickness skin equivalent and automated wounding. J. Vis. Exp. 96, e52576 (2015).
Akhmetshina, A. et al. Dual inhibition of c-abl and PDGF receptor signaling by dasatinib and nilotinib for the treatment of dermal fibrosis. FASEB J. 22, 2214–2222 (2008).
Ashcroft, T., Simpson, J. M. & Timbrell, V. Simple method of estimating severity of pulmonary fibrosis on a numerical scale. J. Clin. Pathol. 41, 467–470 (1988).
Scheuer, P. J. Classification of chronic viral hepatitis: a need for reassessment. J. Hepatol. 13, 372–374 (1991).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Jarjour, M. et al. Fate mapping reveals origin and dynamics of lymph node follicular dendritic cells. J. Exp. Med. 211, 1109–1122 (2014).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Zhang, Y. et al. Model-based analysis of ChIP–seq (MACS). Genome Biol. 9, R137 (2008).
Shen, Y. et al. A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 (2012).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
R Core Team. R: A Language and Environment for Statistical Computing. http://www.R-project.org/ (R Foundation for Statistical Computing, Vienna, 2013).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Enright, A. J. et al. MicroRNA targets in Drosophila. Genome Biol. 5, R1 (2003).
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003).
Dees, C. et al. Platelet-derived serotonin links vascular disease and tissue fibrosis. J. Exp. Med. 208, 961–972 (2011).
Hecker, L. et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat. Med. 15, 1077–1081 (2009).
Popov, Y. et al. Tissue transglutaminase does not affect fibrotic matrix stability or regression of liver fibrosis in mice. Gastroenterology 140, 1642–1652 (2011).
DeClerck, Y., Draper, V. & Parkman, R. Clonal analysis of murine graft-vs-host disease. II. Leukokines that stimulate fibroblast proliferation and collagen synthesis in graft-vs. host disease. J. Immunol. 136, 3549–3552 (1986).
Munde, M. et al. Structure-dependent inhibition of the ETS-family transcription factor PU.1 by novel heterocyclic diamidines. Nucleic Acids Res. 42, 1379–1390 (2014).
Burkholder, T., Foltz, C., Karlsson, E., Linton, C. G. & Smith, J. M. Health evaluation of experimental laboratory mice. Curr. Protoc. Mouse Biol. 2, 145–165 (2012).
Acknowledgements
We thank M. Pascual, K. Dreißigacker, K.-T. Yang, R. Kleinlein and M. Spiller-Becker for technical assistance; and R. Palmisano and P. Tripal of the Optical Imaging Center Erlangen (OICE) for helping with confocal images. This work was supported by grants RA 2506/3-1, RA 2506/4-1, DI 1537/9-1, DI 1537/11-1, DE 2414/2-1, AK 144/2-1, DFG FOR2438, SCHE 1583/7-1, SPP1468, CRC1181 (A01, C01) and SFB TR221 (B04) of the German Research Foundation, the Bundesministerium für Bildung und Forschung (BMBF; project Metharthros), the Marie Curie project OSTEOIMMUNE, the TEAM project of the European Union (305549) and the IMI funded project RTCure (115142), Else Kröner-Fresenius-Stiftung 2014_A184, grants A64, D28 and J40 of the IZKF in Erlangen, grant 16-10-05-1-Ramming of the ELAN-Foundation Erlangen, grant 2017.129.1 of the Wilhelm Sander-Stiftung, grant SNF310030_166259 (to O.D.), NIH (R01 A1057459 and AI095282 to M.H.K.; H1.129063 to G.M.K.P.).
Reviewer information
Nature thanks T. Graf, M. Kolb, M. T. Longaker, C. Nerlov and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
T.W., S.R., J.H.W.D. and A.R. designed the study; T.W., S.R., S.U., M.L., A.S., A.E., S.W., A.-E.M., C.-W.C., C.M., E.K., H.P.K., E.P., C. Dees, F.B. and A.R. acquired data; T.W., S.R., S.U., M.L., A.S., S.W., A.-E.M., C.-W.C., E.K., H.P.K., C.B., E.N., A.J., S.G., O.D., G.S., J.H.W.D. and A.R. interpreted data; C.B., C. Daniel, K.G., A.E.K., E.N., M. Stürzl, F.B., M. Sticherling, S.F., A.K., M.H.K., A.J., S.L.N., D.W.B., G.M.K.P., O.D., G.S., J.H.W.D. and A.R. provided materials; T.W., S.R., G.S., J.H.W.D. and A.R. prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 PU.1-expressing fibroblasts control tissue fibrosis.
a–c, Representative images of immunofluorescence (a, b) and confocal (c) microscopy of human skin, lung, liver, kidney and joint biopsy specimens stained for PU.1 (red), P4Hβ (green), CD45 or CD11b (purple), and with DAPI (blue). Tissues were obtained from healthy individuals (n = 5 each), idiopathic pulmonary fibrosis (n = 4), acute asthma (n = 5), alcoholic liver cirrhosis (n = 4), autoimmune hepatitis (n = 4), cirrhotic kidney (n = 4), interstitial nephritis (n = 5), osteoarthritis (OA; n = 5) and rheumatoid arthritis (RA; n = 5). Haematoxylin and eosin (HE)-stained tissue specimens are included. d, Representative immunofluorescence images (n = 4) of explanted fibrotic fibroblasts stained for PU.1 (red) and one of the following markers (green): FAP, CDH11 or MRC2; nuclei were stained with DAPI (blue). e, Semi-quantification of PU.1+ fibroblasts/total P4Hβ+ fibroblasts per HPF. Tissues were obtained from healthy individuals (n = 5 each), patients with systemic sclerosis (n = 10), plaque psoriasis (n = 7), idiopathic pulmonary fibrosis (n = 4), acute asthma (n = 5), alcoholic liver cirrhosis (n = 4), autoimmune hepatitis (n = 4), cirrhotic kidney (n = 4) and interstitial nephritis (n = 5), osteoarthritis (n = 5) and rheumatoid arthritis (n = 5). f, Cell counts and viability of CRISPR–Cas9-mediated PU.1 knockout in human fibrotic fibroblasts compared to unaffected control fibroblasts and fibroblasts treated with 50% DMSO as toxic control (n = 3 each). Cells were counted per HPF. g, Resting fibroblasts co-transfected with different amounts of a SPI1 expression plasmid as indicated (n = 4 each). Cell viability of fibroblasts was determined by CCK-8 toxicity assay. h–k, Relative Col1a1 and Col1a2 mRNA levels, hydroxyproline concentration, myofibroblast counts per HPF and respective histological scores (skin thickness, Ashcroft, Scheuer) in bleomycin-induced skin fibrosis (h; n = 6 per group), bleomycin-induced lung fibrosis (i; n = 6 per group) and CCl4-induced liver fibrosis (j; n = 5 per group) models, as well as a sclerodermatous chronic graft-versus-host disease model (k; n = 6 per group). Data are mean ± s.e.m. of the indicated number of independent experiments. P values were determined by one-way ANOVA with Tukey’s multiple comparison post hoc test.
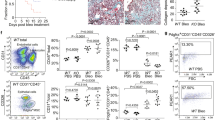
Extended Data Fig. 2 PU.1-expressing fibroblasts in different mouse models of fibrosis.
a, Representative haematoxylin and eosin and immunofluorescence images of a bleomycin-induced skin fibrosis model (n = 8 per group). Mice injected with NaCl were used as controls. b, Mouse model of sclerodermatous chronic graft-versus-host disease (n = 8 per group). Syngeneic transplanted mice were used as controls. c, Fibrosis model of tight skin 1 (Tsk1) mice (n = 11 per group). d, Model of bleomycin-induced pulmonary fibrosis (n = 6 per group). Controls received intratracheal application of NaCl. Representative haematoxylin and eosin and immunofluorescence images of respective tissues stained for PU.1 (red), vimentin (green), and with DAPI (blue) are included. Total Spi1 mRNA in the respective tissues was measured by qPCR. Absolute counts of PU.1-expressing fibroblasts were analysed per HPF. e, f, Mouse model of bleomycin-induced skin fibrosis (n = 5 per group). Controls received NaCl. Representative haematoxylin and eosin and immunofluorescence images of frozen serial tissue sections; boxed areas in the haematoxylin and eosin-stained sections indicate the representative histological regions (yellow, orange and purple) of the correspondingly framed immunofluorescence panels. e, Tissues from control littermates or Spi1GFP reporter mice were stained with DAPI (blue) and the indicated antibodies (red). f, IgG control of NaCl-treated control littermates of Spi1GFP reporter mice (n = 3 per group). g, Semi-quantitative analysis of PU.1 (GFP)-expressing fibroblasts. Absolute counts of PU.1-expressing fibroblasts were analysed per HPF (respective n is given in e). Control images of GFP+ tissue sections are shown in Extended Data Fig. 10d. Data are mean ± s.e.m. of the indicated number of independent experiments. P values were determined by either one-way ANOVA with Tukey’s multiple comparison post hoc test or two-tailed Mann–Whitney U-test if two groups were compared.
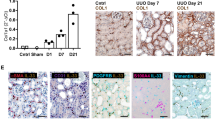
Extended Data Fig. 3 PU.1-expressing fibroblasts in bleomycin-induced lung and CCl4-induced liver fibrosis.
a–e, Mouse model of bleomycin-induced lung fibrosis (n = 4 per group). Controls received NaCl. a, Representative haematoxylin and eosin and immunofluorescence images of frozen serial tissue sections of control littermates or Spi1GFP reporter mice stained with DAPI (blue) and the indicated antibodies (red). b, IgG control of NaCl-treated control littermates of Spi1GFP reporter mice (n = 3 per group). c, Semi-quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 4 each). Absolute counts of PU.1-expressing fibroblasts were analysed per HPF. Control images of GFP+ tissue sections are shown in Extended Data Fig. 10e. d, e, Flow cytometry analysis of digested lungs. d, Gating strategy to characterize GFP+ cells. e, Quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 3 each). The percentage of CD45−vimentin+ PU.1-expressing fibroblasts per lung sample is shown. f–j, Mouse model of CCl4-induced liver fibrosis (n = 4). Controls received oil. f, Representative haematoxylin and eosin and immunofluorescence images of frozen serial tissue sections of control littermates or Spi1GFP reporter mice stained with DAPI (blue) and the indicated antibodies (red). g, IgG control of sunflower oil-treated control littermates of Spi1GFP reporter mice (n = 4 per group). h, Semi-quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 3 each). Absolute counts of PU.1-expressing fibroblasts were analysed per HPF. Control images of GFP+ tissue sections are shown in Extended Data Fig. 10f. i, j, Flow cytometry analysis of digested livers. i, Gating strategy to characterize GFP+ cells. j, Quantitative analysis of PU.1 (GFP)-expressing fibroblasts (n = 4 each). The percentage of CD31−CD45−vimentin+ PU.1-expressing fibroblasts per liver sample is shown. Data are mean ± s.e.m. of the indicated number of biologically independent samples. P values were determined by one-way ANOVA with Tukey’s multiple comparison post hoc test. a, b, f, g, Boxed areas in the haematoxylin and eosin-stained sections indicate the representative histological regions of the corresponding immunofluorescence panels. Experiments were repeated three times independently with similar results.
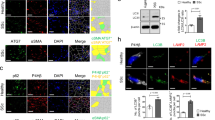
Extended Data Fig. 4 Regulation of PU.1 expression in fibroblasts.
a–c, PU.1 expression levels of primary human fibroblasts. Representative western blot and semi-quantitative analysis of PU.1 protein expression in resting (isolated from normal skin), fibrotic (isolated from fibrotic skin of patients with systemic sclerosis) and inflammatory (isolated from inflamed joints of patients with rheumatoid arthritis) fibroblasts stimulated with or without TNF for 24 h (a), with or without TGF-β for 24 h (b) or for up to 72 h (c) (n = 4 per group). Protein extracts of fibrotic fibroblasts were used as positive control in each lane. d, ChIP analysis (n = 4 each) assessing the binding of SMAD3 to the SPI1 promoter and its −17 kb URE is shown. e, Short interfering RNA (siRNA)-mediated knockdown of SMAD3 in fibrotic fibroblasts stimulated with or without TGF-β for 24 h (n = 5). Scrambled (scr) siRNA was used as control. f, Expression levels of PU.1 in primary human resting, fibrotic and inflammatory fibroblasts (n = 3 each) cultured ex vivo for several passages. g, Expression levels of EZH2 in resting (n = 11), fibrotic (n = 9) and inflammatory (n = 7) fibroblasts relative to β-actin as assessed by western blot analysis. Results are presented relative to resting fibroblasts. h, i, Resting fibroblasts treated with different concentrations of GSK126 as indicated (n = 3 each). h, Cell viability of fibroblasts was determined by CCK-8 toxicity assay. i, Expression levels of H3K27me3 relative to total H3 as assessed by western blot analysis. Results are presented relative to untreated control. j, k, Inflammatory fibroblasts treated with different concentrations of miR-155 antagomirs as indicated (n = 3 each) to investigate cell viability by CCK-8 toxicity assay (j) and mir-155 expression levels relative to let-7b as assessed by qPCR (k). Results are presented relative to cells co-transfected with scrambled antagomirs. l, m, Fibrotic fibroblasts treated with different concentrations of DB1976 to analyse cell viability by CCK-8 toxicity assay (n = 6) (l) and DB1976 dose-dependent effects on TGF-β-induced collagen production (n = 4 each) (m). Results are presented relative to untreated control. Data are mean ± s.e.m. of the indicated number of independent experiments. P values were determined by one-way ANOVA with Tukey’s multiple comparison post hoc test.
Extended Data Fig. 5 Pro-fibrotic potential of PU.1.
a, GSEA of quantitative RNA-seq signals of GO-defined monocyte-related gene clusters in human resting fibroblasts co-transfected with PU.1 (n = 4). Resting fibroblasts co-transfected with a control plasmid were used as controls (n = 4). b, mRNA expression levels of indicated transcripts in human resting fibroblasts treated with or without DB1976 and simultaneously co-transfected with or without a SPI1 expression plasmid (pUNO.1-hSPI1 (PU.1 OE), n = 5 per group) as assessed by qPCR. Cells co-transfected with a scramble (scr) plasmid were used as control. Results are presented relative to cells co-transfected with scramble. c, Genomic annotation of PU.1-binding sites defined by ChIP–seq analysis in primary human fibrotic fibroblasts. d, Annotation of PU.1 ChIP–seq peaks (n = 3 each) at various q-value treshholds to active regulatory elements (AREs). For unbiased identification of active regulatory elements, 11 ENCODE datasets from DNase-sequencing and histone ChIP–seq were used as described in the Methods; q values are those provided by MACS2 call-peak67. e, Differentially expressed genes from gene sets of inflammatory fibroblasts co-transfected with SPI1 (PU.1 OE) or scramble vector as control (ctrl). Gene sets include fibrosis-associated, inflammatory and matrix-degrading pathways determined by qPCR (n = 4 each). Colours represent the significance levels of the observed changes in the respective expression levels in PU.1 overexpression compared to control. f, Micro-mass organoids of inflammatory fibroblasts co-transfected with SPI1 or scramble vector in the presence of TNF for 21 days (n = 8 per group). Sections of micro-mass organoids were stained with haematoxylin and eosin. Lining fibroblasts were quantified relative to total number of cells per HPF. g, Three-dimensional full-thickness skin organoid model of inflammatory fibroblasts co-transfected with SPI1 or scramble vector. The collagen content was measured by hydroxyproline assay; α-SMA expression and skin thickness were quantified per HPF (n = 4 each). h, mRNA expression levels of indicated transcripts in primary human inflammatory fibroblasts treated with or without DB1976 and simultaneously co-transfected with or without miR-155 antagomirs (n = 4 each). Results are presented relative to cells co-transfected with scramble (scr) antagomirs. Data are mean ± s.e.m. of the indicated number of independent experiments. P values were determined either according to a previous study51 (a), by one-way ANOVA with Tukey’s multiple comparison post hoc test (b, h) or by two-tailed Mann–Whitney U-test (e–g).
Extended Data Fig. 6 PU.1 anchors differentiation towards fibrotic fibroblasts in a network of flanking factors including TEAD1.
a, TEAD1 expression levels of primary human fibroblasts. Representative western blot and semi-quantitative analysis of TEAD1 expression in resting, fibrotic and inflammatory fibroblasts (n = 4 each). b, ChIP analysis of TEAD1 binding at regions of genes with a fibrotic signature in the vicinity of PU.1-binding sites. DNA fragments of human fibrotic fibroblasts were immunoprecipitated with anti-TEAD1 and analysed by qPCR relative to input DNA (n = 4 per group). Results are compared to IgG control. Signature pro-fibrotic genes were screened for PU.1 ChIP–seq peaks and potential flanking TEAD1-binding sites. c, mRNA expression levels of indicated transcripts in primary human inflammatory fibroblasts co-transfected with SPI1 or scramble plasmid (n = 4 each); cells were cultured under neutral conditions (serum-starved medium only) or in the presence of TGF-β (fibrotic) or TNF (inflammatory (inflam)). Results are presented relative to scramble under neutral culture conditions. Data are mean ± s.e.m. of the indicated number of biologically independent samples. P values were determined either by one-way ANOVA with Tukey’s multiple comparison post hoc test or two-tailed Mann–Whitney U-test if two groups were compared.
Extended Data Fig. 7 PU.1 silencing in experimental fibrosis.
a–f, Experimental fibrosis models. Representative images of trichrome-stained (a, b) or Sirius red-stained (c, d) tissue sections, mRNA levels of Col1a1 and Col1a2, hydroxyproline content, myofibroblast counts and respective histological scores (skin thickness, Ashcroft, Scheuer) in mice treated with or without DB1976. Mice treated with NaCl or oil were used as controls. a, b, Bleomycin-induced skin fibrosis model with preventive (a; n = 7) or therapeutic (b; n = 8) treatment; in the latter, regression of pre-established fibrosis was evaluated since mice were challenged with bleomycin for 3 weeks to induce robust skin fibrosis before treatment with DB1976 was initiated, while injections with bleomycin were continued. As an additional control, mice were injected with bleomycin for 3 weeks followed by injections with NaCl for another 3 weeks. c, Bleomycin-induced lung fibrosis model (n = 5). d, CCl4-induced liver fibrosis model (n = 5). Body weights (e) and levels of pain and distress (f) of DB1976-treated mice were monitored every second day (n = 5 each). Mice challenged with subcutaneous injections of bleomycin were used as positive controls. Data are mean ± s.e.m. of the indicated number of biologically independent samples. P values were determined by one-way ANOVA with Tukey’s multiple comparison post hoc test.
Extended Data Fig. 8 Effects of DB1976 in anti-fibrotic concentrations on haematopoietic cells and bone marrow-derived stem cells.
a, c, e, g, i, Flow cytometry gating strategy to identify different peripheral blood cell (a) and splenic cell (c) populations, B cell precursors and mature B cells in the bone marrow (e), T cell precursors and mature T cells in the thymus (g) or bone marrow-derived mesenchymal stem cells (MSCs) and haematopoietic stem cells (HSCs) (i) in mice treated with different concentrations of DB1976 or NaCl (n = 3 each) for 6 weeks. FMO, fluorescence minus one controls. b, White blood cell (WBC) count, red blood cell (RBC) count, numbers of thrombocytes (TBCs) and the T to B cell ratio in the peripheral blood. d, Quantification of splenic monocytes (Mø), macrophages (Mph), dendritic cells (DC) and the T to B cell ratio. f, Frequencies of indicated B cell populations. h, Frequency of indicated thymocyte subsets. DN, double-negative thymocytes (based on the expression of CD25 and CD44); DP, double-positive thymocytes. j, Percentage of Lin−CD29+CD105+ MSCs in the bone marrow, mean fluorescence intensity (MFI) of CD29+ MSCs and CD105+ MSCs and percentage of CD45+CD34+ HSCs in the bone marrow are shown. Data are mean ± s.e.m. of the indicated number of biologically independent samples. P values were determined by one-way ANOVA with Tukey’s multiple comparison post hoc test.
Extended Data Fig. 9 Characterization of cultured fibroblast phenotypes.
a–c, Gating strategy of cultured human resting (a), fibrotic (b) and inflammatory (c) fibroblasts stained for PDGFRα, collagen I and vimentin (fibroblast markers) as well as KRT14, CD31, CD45 and CD326 (also known as EpCAM) (control markers). Respective isotype and corresponding positive controls for KRT14 (human keratinocytes), CD31 (human umbilical vein endothelial cells), CD45 (human peripheral blood mononuclear cells) and EpCAM (human kidney tubular epithelial cells) are included. d, e, Proliferation, migration and invasion of different passages (P3, P5, P8) of human resting, fibrotic and inflammatory fibroblasts (n = 3 each) were assessed using the xCELLigence Real Time Cell Analysis Instrument. Resting fibroblasts cultured in the absence of a gradient of chemoattractants were used as controls. Data are mean ± s.e.m. of the indicated number of biologically independent samples. P values were determined by one-way ANOVA with Tukey’s multiple comparison post hoc test.
Extended Data Fig. 10 Control images of human and mouse tissues.
a–c, Representative haematoxylin and eosin and immunofluorescence images of paraffin-embedded (a–c) human skin, lung, liver, kidney and joint tissues (a) or mouse skin, lung and liver tissues (b) stained with DAPI and Ig controls as indicated (n = 5 each). c, Representative images of haematoxylin and eosin and immunofluorescence images of mouse biopsy specimens (n = 5 each) of fibrotic skin, lung and liver stained with DAPI, and for α-SMA, collagen I and CD31. d–f, Representative haematoxylin and eosin and immunofluorescence images of frozen tissue sections of control littermates or Spi1GFP reporter mice stained with DAPI (blue). d, Mouse model of bleomycin-induced skin fibrosis (n = 5 per group). Controls received NaCl. e, Mouse model of bleomycin-induced lung fibrosis (n = 4 per group). Controls received NaCl. f, Mouse model of CCl4-induced liver fibrosis (n = 4 per group). Controls received oil.
Supplementary information
Supplementary Information
This file contains the uncropped blots with size marker indications and Supplementary Table 1, a list of primers used for gene expression analysis.
Source data
Rights and permissions
About this article
Cite this article
Wohlfahrt, T., Rauber, S., Uebe, S. et al. PU.1 controls fibroblast polarization and tissue fibrosis. Nature 566, 344–349 (2019). https://doi.org/10.1038/s41586-019-0896-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-0896-x
This article is cited by
-
Airway and parenchyma transcriptomics in a house dust mite model of experimental asthma
Respiratory Research (2023)
-
Increased synovial immunohistochemistry reactivity of TGF-β1 in erosive peripheral psoriatic arthritis
BMC Musculoskeletal Disorders (2023)
-
Mesenchymal stem cells in fibrotic diseases—the two sides of the same coin
Acta Pharmacologica Sinica (2023)
-
Treatable Traits in Systemic Sclerosis
Clinical Reviews in Allergy & Immunology (2023)
-
Substrate Stiffness and Topography Affect the Morphology of Human Fibroblasts in Mechanical Microenvironment
Journal of Shanghai Jiaotong University (Science) (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.