Abstract
Successful T cell immunotherapy for brain cancer requires that the T cells can access tumour tissues, but this has been difficult to achieve. Here we show that, in contrast to inflammatory brain diseases such as multiple sclerosis, where endothelial cells upregulate ICAM1 and VCAM1 to guide the extravasation of pro-inflammatory cells, cancer endothelium downregulates these molecules to evade immune recognition. By contrast, we found that cancer endothelium upregulates activated leukocyte cell adhesion molecule (ALCAM), which allowed us to overcome this immune-evasion mechanism by creating an ALCAM-restricted homing system (HS). We re-engineered the natural ligand of ALCAM, CD6, in a manner that triggers initial anchorage of T cells to ALCAM and conditionally mediates a secondary wave of adhesion by sensitizing T cells to low-level ICAM1 on the cancer endothelium, thereby creating the adhesion forces necessary to capture T cells from the bloodstream. Cytotoxic HS T cells robustly infiltrated brain cancers after intravenous injection and exhibited potent antitumour activity. We have therefore developed a molecule that targets the delivery of T cells to brain cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All relevant data are included in the manuscript linked as source data; more details are available from the corresponding author on reasonable request
Change history
25 October 2018
Editor’s Note: We would like to alert readers that the reliability of data presented in this manuscript has been the subject of criticisms, which we are currently considering. We will publish an update once our investigation is complete.
20 February 2019
This Article has been retracted; see accompanying Retraction.
References
Davenport, M. P., Grimm, M. C. & Lloyd, A. R. A homing selection hypothesis for T-cell trafficking. Immunol. Today 21, 315–317 (2000).
Krummel, M. F., Bartumeus, F. & Gérard, A. T cell migration, search strategies and mechanisms. Nat. Rev. Immunol. 16, 193–201 (2016).
Carrithers, M. D., Visintin, I., Kang, S. J. & Janeway, C. A., Jr. Differential adhesion molecule requirements for immune surveillance and inflammatory recruitment. Brain 123, 1092–1101 (2000).
Arima, Y. et al. Regulation of immune cell infiltration into the CNS by regional neural inputs explained by the gate theory. Mediators Inflamm. 2013, 898165 (2013).
Sackstein, R., Schatton, T. & Barthel, S. R. T-lymphocyte homing: an underappreciated yet critical hurdle for successful cancer immunotherapy. Lab. Invest. 97, 669–697 (2017).
Fukumura, D., Kloepper, J., Amoozgar, Z., Duda, D. G. & Jain, R. K. Enhancing cancer immunotherapy using antiangiogenics: opportunities and challenges. Nat. Rev. Clin. Oncol. 15, 325–340 (2018).
Cayrol, R. et al. Activated leukocyte cell adhesion molecule promotes leukocyte trafficking into the central nervous system. Nat. Immunol. 9, 137–145 (2008).
Nelissen, J. M. D. T., Peters, I. M., de Grooth, B. G., van Kooyk, Y. & Figdor, C. G. Dynamic regulation of activated leukocyte cell adhesion molecule-mediated homotypic cell adhesion through the actin cytoskeleton. Mol. Biol. Cell 11, 2057–2068 (2000).
Brown, M. H. CD6 as a cell surface receptor and as a target for regulating immune responses. Curr. Drug Targets 17, 619–629 (2016).
Chappell, P. E. et al. Structures of CD6 and its ligand CD166 give insight into their interaction. Structure 23, 1426–1436 (2015).
Li, Y. et al. CD6 as a potential target for treating multiple sclerosis. Proc. Natl Acad. Sci. USA 114, 2687–2692 (2017).
Bullard, D. C. et al. Intercellular adhesion molecule-1 expression is required on multiple cell types for the development of experimental autoimmune encephalomyelitis. J. Immunol. 178, 851–857 (2007).
Kijima, N. et al. CD166/activated leukocyte cell adhesion molecule is expressed on glioblastoma progenitor cells and involved in the regulation of tumor cell invasion. Neuro-oncol. 14, 1254–1264 (2012).
Rosenberg, S. A. & Restifo, N. P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015).
Bonini, C. & Mondino, A. Adoptive T-cell therapy for cancer: The era of engineered T cells. Eur. J. Immunol. 45, 2457–2469 (2015).
Hansen, A. G. et al. ALCAM/CD166 is a TGF-β-responsive marker and functional regulator of prostate cancer metastasis to bone. Cancer Res. 74, 1404–1415 (2014).
Peñuelas, S. et al. TGF-β increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15, 315–327 (2009).
Gu, M.-X. et al. Proteomic analysis of endothelial lipid rafts reveals a novel role of statins in antioxidation. J. Proteome Res. 11, 2365–2373 (2012).
Dorovini-Zis, K. The Blood–Brain Barrier in Health and Disease, Volume One: Morphology, Biology and Immune Function (CRC Press, London, 2015).
Carman, C. V. & Springer, T. A. A transmigratory cup in leukocyte diapedesis both through individual vascular endothelial cells and between them. J. Cell Biol. 167, 377–388 (2004).
Muller, W. A. Mechanisms of leukocyte transendothelial migration. Annu. Rev. Pathol. 6, 323–344 (2011).
McEver, R. P. & Zhu, C. Rolling cell adhesion. Annu. Rev. Cell Dev. Biol. 26, 363–396 (2010).
Engelhardt, B. Molecular mechanisms involved in T cell migration across the blood-brain barrier. J. Neural Transm. (Vienna) 113, 477–485 (2006).
Laschinger, M., Vajkoczy, P. & Engelhardt, B. Encephalitogenic T cells use LFA-1 for transendothelial migration but not during capture and initial adhesion strengthening in healthy spinal cord microvessels in vivo. Eur. J. Immunol. 32, 3598–3606 (2002).
Green, C. E. et al. Dynamic shifts in LFA-1 affinity regulate neutrophil rolling, arrest, and transmigration on inflamed endothelium. Blood 107, 2101–2111 (2006).
Orta-Mascaró, M. et al. CD6 modulates thymocyte selection and peripheral T cell homeostasis. J. Exp. Med. 213, 1387–1397 (2016).
Calderwood, D. A. & Ginsberg, M. H. Talin forges the links between integrins and actin. Nat. Cell Biol. 5, 694–697 (2003).
Lawson, C. et al. FAK promotes recruitment of talin to nascent adhesions to control cell motility. J. Cell Biol. 196, 223–232 (2012).
Poria, R. B. et al. Characterization of a radiolabeled small molecule targeting leukocyte function-associated antigen-1 expression in lymphoma and leukemia. Cancer Biother. Radiopharm. 21, 418–426 (2006).
Baumann, K. Cell adhesion: FAK or talin: who goes first? Nat. Rev. Mol. Cell Biol. 13, 138–139 (2012).
Critchley, D. R. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem. Soc. Trans. 32, 831–836 (2004).
Mitra, S. K., Hanson, D. A. & Schlaepfer, D. D. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 (2005).
Cavalcanti-Adam, E. A. et al. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 92, 2964–2974 (2007).
Hegde, M. et al. Tandem CAR T cells targeting HER2 and IL13Rα2 mitigate tumor antigen escape. J. Clin. Invest. 126, 3036–3052 (2016).
Liu, Y. et al. Regulation of leukocyte transmigration: cell surface interactions and signaling events. J. Immunol. 172, 7–13 (2004).
Auerbach, S. D., Yang, L. & Luscinskas, F. W. in Adhesion Molecules: Function and Inhibition 99–116 (Springer, Basel, 2007).
Steiner, O. et al. Differential roles for endothelial ICAM-1, ICAM-2, and VCAM-1 in shear-resistant T cell arrest, polarization, and directed crawling on blood-brain barrier endothelium. J. Immunol. 185, 4846–4855 (2010).
Lee, B. P. L. & Imhof, B. A. Lymphocyte transmigration in the brain: a new way of thinking. Nat. Immunol. 9, 117–118 (2008).
Bughani, U. et al. T cell activation and differentiation is modulated by a CD6 domain 1 antibody Itolizumab. PLoS One 12, e0180088 (2017).
Ager, A., Watson, H. A., Wehenkel, S. C. & Mohammed, R. N. Homing to solid cancers: a vascular checkpoint in adoptive cell therapy using CAR T-cells. Biochem. Soc. Trans. 44, 377–385 (2016).
D’Aloia, M. M., Zizzari, I. G., Sacchetti, B., Pierelli, L. & Alimandi, M. CAR-T cells: the long and winding road to solid tumors. Cell Death Dis. 9, 282 (2018).
Ahmed, N. et al. HER2-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: a phase 1 dose-escalation trial. JAMA Oncol. 3, 1094–1101 (2017).
Manders, E. M. M., Verbeek, F. J. & Aten, J. A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 169, 375–382 (1993).
Ahmed, N. et al. HER2-specific T cells target primary glioblastoma stem cells and induce regression of autologous experimental tumors. Clin. Cancer Res. 16, 474–485 (2010).
Sudha, T. et al. Nanoparticulate tetrac inhibits growth and vascularity of glioblastoma xenografts. Horm. Cancer 8, 157–165 (2017).
Paris, D. et al. Impaired orthotopic glioma growth and vascularization in transgenic mouse models of Alzheimer’s disease. J. Neurosci. 30, 11251–11258 (2010).
Acknowledgements
We thank M. K. Brenner and C. Gillespie for scientific advice and linguistic editing, respectively, and S. Roberge and M. Duquette for technical assistance. The D3 antibody was a gift from M. Brown. This work was funded by an SU2C-St. Baldrick’s Pediatric Dream Team Translational Research Grant (SU2C-AACR-DT1113; NA/PS/MDT). SU2C is a program of the Entertainment Industry Foundation administered by the American Association for Cancer Research. Also funded by Alex’s Lemonade Stand, NIH-T32HL092332 (K.F./T.B.; H. Heslop), R01AI067946 (J.S.O.), T32GM08812 (K.F./T.B.; M. Estes), DK56338, CA125123, and 1S10OD020151-01, CPRIT (RP150578), the Dan L. Duncan CCC and P01-CA080124 (R.K.J./D.F.); R35-CA197743, K08-GM123261 (F.L.) and P50-CA165962 (R.K.J.). This content does not necessarily represent the official views of the funding agencies, the Department of Veterans Affairs or the U.S. Government.
Reviewer information
Nature thanks M. Platten and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
N.A. conceived the main study idea, and with H.S. conceived and implemented the study details. N.A., K.F. and A.P. designed HS molecules. H.S., M.D.T., S.P.M., P.S., S.E.-N., M.H., F.S., J.D. and N.A. performed the CAM studies. M.L.B. performed in silico modelling. F.L. and H.S. designed and implemented microfluidic experiments. M.Ma., T.B., S.K.J. and A.Z.G. performed molecular testing. H.S., F.S., J.D., M.Mu. and J.S.O. designed and performed the subcellular imaging experiments. J.R., H.S., V.S.S., A.S., T.S., S.P.M., S.-H.H., D.F., S.K., R.K.J. and N.A. implemented the animal microscopy and experiments. All authors gave final approval.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
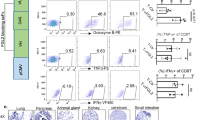
Extended Data Fig. 1 Analysis of CAM expression in primary brain tumours.
a, High-throughput IFC analysis of the endothelial adhesion molecules ICAM1, VCAM1, and ALCAM in 93 primary GBM, 25 primary medulloblastoma and 5 normal brain samples. MATLAB segmentation and masking analysis algorithm of the co-immunofluorescence (co-IFC) of ICAM1 or VCAM (acquired on 647 channel), CD31 (acquired on 594 channel) and ALCAM (acquired on 488 channel), and DAPI (acquired on blue/cyan channel). b, Isolation and characterization of pTECs. Flow cytometry sorting gating strategy of pTECs from freshly excised glioblastoma (GBM; n = 5) based on CD31 positivity. Isolated GBM endothelial cells also expressed the endothelial markers VE-cadherin, von Willebrand Factor (vWF) and ALCAM. Isotype shown in lighter grey and test shown in darker grey in individual histograms. n = 5 surgical samples each interrogated at least twice. At least 100,000 events were acquired per condition.
Extended Data Fig. 2 ALCAM expression in a panel of human and mouse endothelial cells and their reactivity to inflammatory and cancerous conditioning.
a, Western blot for ALCAM in a panel of human and mouse endothelial cell lines: pTECs, HBMECs, 1ry BMECs, 1ry PVECs, HUVECs, HMVEC-Ls, bEnd.3 (mouse brain tumour EC) and 2-H11 (mouse SV40-transformed axillary lymph node vascular endothelium). Left, basal ALCAM expression except in tumour endothelial cells (pTEC and 2-H11). Right, induction of ALCAMs in all endothelial cells after incubation with TNFα for 6 h. b, Expression of ALCAM at baseline and after 6 h of conditioning in GBM supernatant, TGFβ or IL6. Only tumour endothelial cells expressed ALCAM at baseline while normal endothelial cells did not. c, IFC for ALCAM in 5 × 104 pTECs and HBMECs in the in vitro BBB model at baseline and after culture in GBM supernatant. Scale bars, 50 μm. d, Differential expression of key adhesion molecules at baseline and under the influence of cancer and inflammation in pTECs and HBMECs. Flow cytometry dot plots detailing baseline expression of ALCAM, VCAM1 and ICAM1 on 1 × 104 pTECs and HBMECs and conditioned expression after culture in GBM supernatant, TGFβ or IL6. e–h, Expression of adhesion molecules at baseline and under the influence of cancer and inflammation in pTECs (n = 4) acquired from surgically resected samples (pTEC #1 is shown in Fig. 1g).
Extended Data Fig. 3 In silico design of the prototype and derivative HS molecules, their forced expression and detection on T cells, and studies of their in vitro dynamic interactions with endothelial cells under shear stress.
a, The potential interaction between ALCAM V1 (grey ribbon) and CD6 from computational docking. D1 of CD6 is coloured blue, D2 green and D3 orange. b, Details of the potential interaction interface between ALCAM V1 (grey ribbon) and CD6 D3 (orange ribbon). A rendering of the electrostatic surface of ALCAM V1 (grey ribbon) with the D3 domain of CD6 (orange ribbon) in the same orientation. Potential interacting residues are highlighted in the models and in a diagram generated from PDBe PISA and PDBSum (c). A small region of positively charged residues in ALCAM V1 appears to interact with a negatively charged patch of residues on CD6 D3. d, Structure of the prototype HS molecule. e, HS multimers 3HS and 5HS. f, HS molecules with non-signalling endodomains, HSΔ, 3HSΔ and 5HSΔ. g, Strategy used for surface detection of the HS exodomain using a D3-specific antibody and specific binding of the HS exodomain to soluble ALCAM. h, Flow cytometry confirming HS surface expression (using D3 monoclonal antibody) on T cells. i, Design of the HS–ALCAM PLA experiment. j, Digital rendition of PLA using ImageTool. The ALCAM probe (–) binds to the D3 probe (+) to trigger the PCR generating the red fluorescent signal that is quantified as total signal per region (TSR) in Fig. 2f. k, l, Dynamic microfluidic studies showing still image from Supplementary Video 1 of Bioflux channels with non-transduced control (NT) T cells (top) versus 1 × 106 HS T cells interrogated under shear force over an ALCAM-expressing endothelium (k), and still image from MJtracker demonstrating various T cells under interrogation for various TEM dynamic measures, the standard grid used and the equation used for calculations (l). m, Dynamic adhesion of T cells to endothelial cells per field of view. n, Average dynamic rolling velocity against time; *P < 0.05, **P < 0.01, ***P < 0.001. Two-way ANOVA with Tukey’s test for multiple-comparisons (compared to NT cells).
Extended Data Fig. 4 Functional effects of elimination of ALCAM on endothelial cells and knockout of human ALCAM using CRISPR–Cas9 technology and its effect on T cell migration across the BBB.
a, Flow cytometry of ALCAM expression on 1 × 106 wild-type pTECs at base-line, after TGFβ induction of ALCAM and after being transfected with 25 nM ALCAM siRNA for 48 h to knock down (KD) ALCAM. Transmigration assay using pTECs to simulate a cancerous BBB showing percentage of migrant T cells compared with ALCAM-KD is shown in Fig. 2k. b, Highest three scoring guide RNA designs (sgRNA-44, sgRNA-45 and sgRNA-49) as seen on the SnapGene software intended to disrupt ALCAM exons for the extracellular and transmembrane moiety. c, d, CD5-KO (c) and CD19-KO (d) were used as positive and negative experimental controls, respectively. e, Flow cytometry of ALCAM expression on wild-type HUVECs and HUVEC ALCAM-KO using CRISPR–Cas9 (using the guide sgRNA-45) assessed at baseline and after TGFβ incubation. Isotype was used as control. f, Transmigration assay showing percentage of 2 × 106 migrating T cells on wild-type HUVECs before and after ALCAM induction compared to ALCAM-KO HUVECs. Both experiments were done at baseline then after ALCAM induction was confirmed. Data shown as mean ± s.d. (n ≥ 3 experiments; donor T cells n = 3), **P < 0.01, ***P < 0.001. Tukey’s test (compared to wild-type pTECs). g, RT–PCR analysis of representative of 1 × 106 ALCAM-KO HS T cells in comparison to wild-type normal T cells. GAPDH was used as an internal control. h, Flow cytometry showing >90% knockout efficiency of the three sgRNAs on 1 × 105 T cells in comparison to wild-type normal T cells; CRISPR–Cas9 only and isotype were used as experimental controls. i, Sorted ALCAM-negative KO T cells were then successfully transduced with the six HS constructs. j, Transmigration assay showing percentage of 2 × 105 migrating T cells on a cBBB model to compare wild-type with ALCAM-KO T cells in four conditions (ALCAM–, ALCAM+ conditioned with TGFβ, after blocking ALCAM, after washing the blocking away). Data in f and j are shown as mean ± s.d. (n ≥ 3 experiments; donors n = 3), *P < 0.05, **P < 0.01. Tukey’s test (compared to ALCAM+ T cells).
Extended Data Fig. 5 Flow cytometry quantification of nodes downstream of CD6 signalling endodomains and high-throughput analysis of super-resolution imaging using deconvolution microscopy.
a–c, Quantification of the flow cytometric data for LFA-1 open configuration (a), pZap70 (b) and talin (c) before (solid bars) and after (dotted bars) TWM of 1 × 105 T cells. ***P < 0.001. d, Characterization of cellular features of migrant T cells using collective quantification of actin MFI, focal adhesions at HS–ALCAM interface, area of spreading, and podosynapse formation by high-throughput microscopy in three donors. n = 200–800 cells. e, Box plot summary representing single-cell data distributions of all replicates between all three donors expressing HS versus NT controls. Centre lines, data median. Boxes, middle quartiles. Whiskers, upper and lower limits.
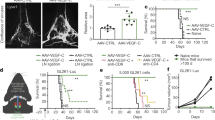
Extended Data Fig. 6 Assessment of TILs in GBM explants.
a, Flow cytometry of 1 × 104 TILs; all HS T cell designs compared with normal T cells gated on CD3+CD45+ then D3+ fractions in GBM explants 24 h after intravenous infusion. Representative plots shown. n = 5 animals per group. b, Cranial window on a live mouse bearing U87-GBM tumour (black arrow, right).
Extended Data Fig. 7 Analysis of T cell infiltrates in vital organs and normal brain after infusion of HS T cells.
a, CD3 immunohistochemistry (IHC) staining of normal vital tissues from animals receiving HS T cells or NT control cells. n = 3 mice per group. Scale bars, 40 μm. b, IHC showing HS T cell infiltrate in micro-dissected GBM xenograft. Scoring of CD3+ DAB signal was analysed using IHC-Profiler plugin in ImageJ. Respective image analysis output and the score assigned using IHC-Profiler are also shown for each image. Total percentage of CD3+ DAB signal was more 66% in all mouse brain with HS T cells (scores 3–4) and percentages in control mice were less than 20% (scores 0–1). Scale bars, 50 μm. n = 3 mice per group.
Extended Data Fig. 8 Characterization of therapeutic T cells after transmigration through an in vitro BBB model.
a, Flow cytometry assessing HER2-CAR and HS molecule expression in HS HER2-CAR T cells. b–d, 1 × 105 T cells were collected from the bottom chamber after transmigration on ALCAM-expressing endothelium and analysed for CD45RO and CCR7 to assess their centrality (b), expression of the exhaustion markers PD-1 (black), TIM-3 (red) and LAG3 (orange) (before transmigration is shown in grey) (c), and proliferative capacity before (red) and after (blue) transmigration, using efLuor 670 (d).
Extended Data Fig. 9 Analysis of TILs isolated from tumour xenografts and normal brain for HER2-CAR HS T cells.
a, Flow cytometry of TILs isolated from orthotopic tumour xenografts 24 h after intravenous injection of HS T cell products, HER2-CAR T cells and NT control T cells. Xenografts were micro-dissected and TILs were isolated and enriched on a percoll/ficoll gradient. Cells were gated on D3+ subset inside a gate of D3+CD45+. A subset of HER2-CAR inside a gate of CD3+CD45+D3+ was used to detect HER2-CAR HS T cells specifically. n = 5 mice per group, representative data shown. b, Flow cytometry following the same gating strategy indicating the absence of HS T cells in the contralateral lobe to the tumour xenograft; data representative of three mice.
Extended Data Fig. 10 Overexpression of full-length native CD6 and its phenotypic and functional effects on T cells.
a, Cloning strategy of native CD6 in an SFG retroviral backbone. b, Flow cytometry showing the transduction of 1 × 105 native CD6 relative to HS constructs on T cells. c, Flow cytometry of the activation marker CD69 on day 8 after transduction without additional stimulation. d, Flow cytometry of the activation and exhaustion marker PD-1 stained with PD-1 PerCP on day 8 transduction at basal level without additional stimulation. e, Expansion plot of T cells expressing the native CD6 relative to NT and various HS T cells; cells were grown in IL-7/IL-15 and collected at day 2 and day 12 post transduction. f, Transmigration of 2 × 105 T cells through a cancerous BBB model showing the percentage of migrant T cells expressing native CD6 relative to various HS T cells, and the response to blocking ALCAM and its restitution. Data shown as mean ± s.d. (n ≥ 3 experiments; donor T cells, n = 3) ***P < 0.001 compared to migration of CD6 through ALCAM+ BBB. ANOVA with Tukey’s post-hoc analysis.
Supplementary information
Supplementary Methods
This file contains Supplementary Methods and additional references.
Supplementary Video 1
Microfluidics studies of HS and NT T-cells.
Supplementary Video 2
Representative flowing and adherence of 5HS T-cells in the brain tumour vascular bed.
Supplementary Video 3
Representative rolling and extravasation of 5HS T-cells in the brain tumour vascular bed.
Supplementary Video 4
Representative flowing and adherence of NT T-cells in the brain tumour vascular bed.
Supplementary Video 5
Representative rolling of NT T-cells in the brain tumour vascular bed.
Source data
Rights and permissions
About this article
Cite this article
Samaha, H., Pignata, A., Fousek, K. et al. A homing system targets therapeutic T cells to brain cancer. Nature 561, 331–337 (2018). https://doi.org/10.1038/s41586-018-0499-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-018-0499-y
Keywords
This article is cited by
-
Strategies to package recombinant Adeno-Associated Virus expressing the N-terminal gasdermin domain for tumor treatment
Nature Communications (2021)
-
Modulation of cell adhesion and migration through regulation of the immunoglobulin superfamily member ALCAM/CD166
Clinical & Experimental Metastasis (2019)
-
T cells engineered to home in on brain cancer
Nature (2018)
-
T cells home in on brain cancer
Nature Reviews Immunology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.