Abstract
Viral infection is a major contributor to the global cancer burden. Recent advances have revealed that seven known oncogenic viruses promote tumorigenesis through shared host cell targets and pathways. A comprehensive understanding of the principles of viral oncogenesis may enable the identification of unknown infectious aetiologies of cancer and the development of therapeutic or preventive strategies for virus-associated cancers. In this Review, we discuss the molecular mechanisms of viral oncogenesis in humans. We highlight recent advances in understanding how viral manipulation of host cellular signalling, DNA damage responses, immunity and microRNA targets promotes the initiation and development of cancer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
zur Hausen, H. & de Villiers, E. M. Cancer “causation” by infections — individual contributions and synergistic networks. Semin. Oncol. 41, 860–875 (2014).
Moore, P. S. & Chang, Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer 10, 878–889 (2010).
Epstein, M. A., Achong, B. G. & Barr, Y. M. Virus particles in cultured lymphoblasts from Burkitt’s lymphoma. Lancet 1, 702–703 (1964).
Chang, Y. et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869 (1994). In this study, representational difference analysis identifies KSHV as a new human herpesvirus associated with Kaposi sarcoma.
Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 110, 525–541 (2006).
Liu, W. et al. Identifying the target cells and mechanisms of Merkel cell polyomavirus infection. Cell Host Microbe 19, 775–787 (2016). This study reveals that human dermal fibroblasts are natural host cells of MCPyV infection in human skin.
Feng, H., Shuda, M., Chang, Y. & Moore, P. S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319, 1096–1100 (2008). This study discovers that clonal integration of MCPyV DNA into the genome of Merkel cell carcinoma is a key event for tumour development.
Durst, M., Gissmann, L., Ikenberg, H. & zur Hausen, H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc. Natl Acad. Sci. USA 80, 3812–3815 (1983).
Choo, Q. L. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 (1989).
Blumberg, B. S., Alter, H. J. & Visnich, S. A “new” antigen in leukemia sera. JAMA 191, 541–546 (1965).
Poiesz, B. J. et al. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T cell lymphoma. Proc. Natl Acad. Sci. USA 77, 7415–7419 (1980).
Sherr, C. J. & McCormick, F. The RB and p53 pathways in cancer. Cancer Cell 2, 103–112 (2002).
McLaughlin-Drubin, M. E. & Munger, K. Viruses associated with human cancer. Biochim. Biophys. Acta 1782, 127–150 (2008).
Howley, P. M. & Livingston, D. M. Small DNA tumor viruses: large contributors to biomedical sciences. Virology 384, 256–259 (2009).
Wright, D. G. et al. Human T cell leukemia virus type-1-encoded protein HBZ represses p53 function by inhibiting the acetyltransferase activity of p300/CBP and HBO1. Oncotarget 7, 1687–1706 (2016).
Ringelhan, M., O’Connor, T., Protzer, U. & Heikenwalder, M. The direct and indirect roles of HBV in liver cancer: prospective markers for HCC screening and potential therapeutic targets. J. Pathol. 235, 355–367 (2015).
Taylor, J. M. & Nicot, C. HTLV-1 and apoptosis: role in cellular transformation and recent advances in therapeutic approaches. Apoptosis 13, 733–747 (2008).
Morales-Sanchez, A. & Fuentes-Panana, E. M. Human viruses and cancer. Viruses 6, 4047–4079 (2014).
Liu, W., MacDonald, M. & You, J. Merkel cell polyomavirus infection and Merkel cell carcinoma. Curr. Opin. Virol. 20, 20–27 (2016).
Knoll, S. et al. Dissection of cell context-dependent interactions between HBx and p53 family members in regulation of apoptosis: a role for HBV-induced HCC. Cell Cycle 10, 3554–3565 (2011).
Cho, D. C. Targeting the PI3K/Akt/mTOR pathway in malignancy: rationale and clinical outlook. BioDrugs 28, 373–381 (2014).
Wong, J. P. & Damania, B. Modulation of oncogenic signaling networks by Kaposi’s sarcoma-associated herpesvirus. Biol. Chem. 398, 911–918 (2017).
Noch, E. & Khalili, K. Oncogenic viruses and tumor glucose metabolism: like kids in a candy store. Mol. Cancer Ther. 11, 14–23 (2012).
Surviladze, Z., Sterk, R. T., DeHaro, S. A. & Ozbun, M. A. Cellular entry of human papillomavirus type 16 involves activation of the phosphatidylinositol 3-kinase/Akt/mTOR pathway and inhibition of autophagy. J. Virol. 87, 2508–2517-(2013).
Zhang, L., Wu, J., Ling, M. T., Zhao, L. & Zhao, K. N. The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol. Cancer 14, 87 (2015).
Fukuda, M. & Longnecker, R. Latent membrane protein 2A inhibits transforming growth factor-beta 1-induced apoptosis through the phosphatidylinositol 3-kinase/Akt pathway. J. Virol. 78, 1697–1705 (2004).
Scholle, F., Bendt, K. M. & Raab-Traub, N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J. Virol. 74, 10681–10689 (2000).
Olagnier, D. et al. HTLV-1 Tax-mediated inhibition of FOXO3a activity is critical for the persistence of terminally differentiated CD4+ T cells. PLoS Pathog. 10, e1004575 (2014).
Stallone, G. et al. Sirolimus for Kaposi’s sarcoma in renal-transplant recipients. N. Engl. J. Med. 352, 1317–1323 (2005).
Chang, H. H. & Ganem, D. A unique herpesviral transcriptional program in KSHV-infected lymphatic endothelial cells leads to mTORC1 activation and rapamycin sensitivity. Cell Host Microbe 13, 429–440 (2013).
Sodhi, A. et al. Akt plays a central role in sarcomagenesis induced by Kaposi’s sarcoma herpesvirus-encoded G protein-coupled receptor. Proc. Natl Acad. Sci. USA 101, 4821–4826 (2004).
Tomlinson, C. C. & Damania, B. The K1 protein of Kaposi’s sarcoma-associated herpesvirus activates the Akt signaling pathway. J. Virol. 78, 1918–1927 (2004).
Shuda, M., Kwun, H. J., Feng, H., Chang, Y. & Moore, P. S. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J. Clin. Invest. 121, 3623–3634 (2011).
Kim, E. K. & Choi, E. J. Compromised MAPK signaling in human diseases: an update. Arch. Toxicol. 89, 867–882 (2015).
Menzel, N. et al. MAP-kinase regulated cytosolic phospholipase A2 activity is essential for production of infectious hepatitis C virus particles. PLoS Pathog. 8, e1002829 (2012).
Bowser, B. S., Alam, S. & Meyers, C. Treatment of a human papillomavirus type 31b-positive cell line with benzo[a]pyrene increases viral titer through activation of the Erk1/2 signaling pathway. J. Virol. 85, 4982–4992 (2011).
Matusali, G., Arena, G., De Leo, A., Di Renzo, L. & Mattia, E. Inhibition of p38 MAP kinase pathway induces apoptosis and prevents Epstein Barr virus reactivation in Raji cells exposed to lytic cycle inducing compounds. Mol. Cancer 8, 18 (2009).
Dawson, C. W., Laverick, L., Morris, M. A., Tramoutanis, G. & Young, L. S. Epstein-Barr virus-encoded LMP1 regulates epithelial cell motility and invasion via the ERK-MAPK pathway. J. Virol. 82, 3654–3664 (2008).
McCormick, C. & Ganem, D. The kaposin B protein of KSHV activates the p38/MK2 pathway and stabilizes cytokine mRNAs. Science 307, 739–741 (2005).
Xia, L. et al. Upregulated FoxM1 expression induced by hepatitis B virus X protein promotes tumor metastasis and indicates poor prognosis in hepatitis B virus-related hepatocellular carcinoma. J. Hepatol. 57, 600–612 (2012).
Ranganathan, P., Weaver, K. L. & Capobianco, A. J. Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11, 338–351 (2011).
South, A. P., Cho, R. J. & Aster, J. C. The double-edged sword of Notch signaling in cancer. Semin. Cell Dev. Biol. 23, 458–464 (2012).
Rozenblatt-Rosen, O. et al. Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487, 491–495 (2012). This study analyses the host interactome and transcriptome networks perturbed by DNA tumour virus proteins, revealing cancer driver genes that contribute to the molecular basis of human cancer.
Tan, M. J. et al. Cutaneous β-human papillomavirus E6 proteins bind Mastermind-like coactivators and repress Notch signaling. Proc. Natl Acad. Sci. USA 109, E1473–E1480 (2012).
Brimer, N., Lyons, C., Wallberg, A. E. & Vande Pol, S. B. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene 31, 4639–4646 (2012).
Meyers, J. M., Spangle, J. M. & Munger, K. The human papillomavirus type 8 E6 protein interferes with NOTCH activation during keratinocyte differentiation. J. Virol. 87, 4762–4767 (2013).
Rowe, M., Raithatha, S. & Shannon-Lowe, C. Counteracting effects of cellular Notch and Epstein-Barr virus EBNA2: implications for stromal effects on virus-host interactions. J. Virol. 88, 12065–12076 (2014).
Gao, J. et al. Hepatitis B virus X protein activates Notch signaling by its effects on Notch1 and Notch4 in human hepatocellular carcinoma. Int. J. Oncol. 48, 329–337 (2016).
Curry, C. L. et al. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi’s sarcoma tumor cells. Oncogene 24, 6333–6344 (2005).
Emuss, V. et al. KSHV manipulates Notch signaling by DLL4 and JAG1 to alter cell cycle genes in lymphatic endothelia. PLoS Pathog. 5, e1000616 (2009).
Liu, R. et al. KSHV-induced notch components render endothelial and mural cell characteristics and cell survival. Blood 115, 887–895 (2010).
Cheng, F. et al. KSHV-initiated notch activation leads to membrane-type-1 matrix metalloproteinase-dependent lymphatic endothelial-to-mesenchymal transition. Cell Host Microbe 10, 577–590 (2011). In this study, KSHV is found to activate the Notch signalling pathway to induce lymphatic endothelial cells to mesenchymal cell transition, which may contribute to Kaposi sarcoma development by promoting the generation of infected cells with invasive mesenchymal cell properties.
Lan, K. et al. Kaposi’s sarcoma herpesvirus-encoded latency-associated nuclear antigen stabilizes intracellular activated Notch by targeting the Sel10 protein. Proc. Natl Acad. Sci. USA 104, 16287–16292 (2007).
Nusse, R. & Clevers, H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169, 985–999 (2017).
Fujimuro, M. & Hayward, S. D. The latency-associated nuclear antigen of Kaposi’s sarcoma-associated herpesvirus manipulates the activity of glycogen synthase kinase-3β. J. Virol. 77, 8019–8030 (2003).
Morrison, J. A., Gulley, M. L., Pathmanathan, R. & Raab-Traub, N. Differential signaling pathways are activated in the Epstein-Barr virus-associated malignancies nasopharyngeal carcinoma and Hodgkin lymphoma. Cancer Res. 64, 5251–5260 (2004).
Morrison, J. A., Klingelhutz, A. J. & Raab-Traub, N. Epstein-Barr virus latent membrane protein 2A activates β-catenin signaling in epithelial cells. J. Virol. 77, 12276–12284 (2003).
Jha, H. C., Banerjee, S. & Robertson, E. S. The role of gammaherpesviruses in cancer pathogenesis. Pathogens 5, 42 (2016).
Daud, M., Rana, M. A., Husnain, T. & Ijaz, B. Modulation of Wnt signaling pathway by hepatitis B virus. Arch. Virol. 16, 2937–2947 (2017).
Ma, G., Yasunaga, J., Fan, J., Yanagawa, S. & Matsuoka, M. HTLV-1 bZIP factor dysregulates the Wnt pathways to support proliferation and migration of adult T cell leukemia cells. Oncogene 32, 4222–4230 (2013).
Zhang, Q., Lenardo, M. J. & Baltimore, D. 30 Years of NF-κB: a blossoming of relevance to human pathobiology. Cell 168, 37–57 (2017).
Karin, M. Nuclear factor-kappaB in cancer development and progression. Nature 441, 431–436 (2006).
Kulwichit, W. et al. Expression of the Epstein-Barr virus latent membrane protein 1 induces B cell lymphoma in transgenic mice. Proc. Natl Acad. Sci. USA 95, 11963–11968 (1998).
Luftig, M. et al. Epstein-Barr virus latent membrane protein 1 activation of NF-κB through IRAK1 and TRAF6. Proc. Natl Acad. Sci. USA 100, 15595–15600 (2003).
Wang, L. W., Jiang, S. & Gewurz, B. E. Epstein-Barr virus LMP1-mediated oncogenicity. J. Virol. 91, e01718–16 (2017).
Gopalakrishnan, R., Matta, H. & Chaudhary, P. M. A purine scaffold HSP90 inhibitor BIIB021 has selective activity against KSHV-associated primary effusion lymphoma and blocks vFLIP K13-induced NF-κB. Clin. Cancer Res. 19, 5016–5026 (2013).
Bagneris, C. et al. Crystal structure of a vFlip-IKKγ complex: insights into viral activation of the IKK signalosome. Mol. Cell 30, 620–631 (2008).
Chugh, P. et al. Constitutive NF-κB activation, normal Fas-induced apoptosis, and increased incidence of lymphoma in human herpes virus 8 K13 transgenic mice. Proc. Natl Acad. Sci. USA 102, 12885–12890 (2005).
Lavorgna, A. & Harhaj, E. W. Regulation of HTLV-1 tax stability, cellular trafficking and NF-κB activation by the ubiquitin-proteasome pathway. Viruses 6, 3925–3943 (2014).
Ciccia, A. & Elledge, S. J. The DNA damage response: making it safe to play with knives. Mol. Cell 40, 179–204 (2010).
Lilley, C. E., Schwartz, R. a. & Weitzman, M. D. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15, 119–126 (2007).
Weitzman, M. D., Lilley, C. E. & Chaurushiya, M. S. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64, 61–81 (2010).
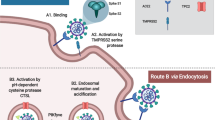
Fradet-Turcotte, A. et al. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J. Virol. 85, 8996–9012 (2011).
Gillespie, K. A., Mehta, K. P., Laimins, L. A. & Moody, C. A. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol. 86, 9520–9526 (2012).
Moody, C. A. & Laimins, L. A. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5, e1000605 (2009).
Sakakibara, N., Mitra, R. & McBride, A. A. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 85, 8981–8995 (2011).
Sowd, G. A., Li, N. Y. & Fanning, E. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog. 9, e1003283 (2013).
Tsang, S. H., Wang, X., Li, J., Buck, C. B. & You, J. Host DNA damage response factors localize to Merkel cell polyomavirus DNA replication sites to support efficient viral DNA replication. J. Virol. 88, 3285–3297 (2014).
Weitzman, M. D. & Weitzman, J. B. What’s the damage? The impact of pathogens on pathways that maintain host genome integrity. Cell Host Microbe 15, 283–294 (2014).
Duensing, S. & Munger, K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 62, 7075–7082 (2002).
Wallace, N. A. et al. High-risk alphapapillomavirus oncogenes impair the homologous recombination pathway. J. Virol. 91, e01084–17 (2017).
Gruhne, B. et al. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc. Natl Acad. Sci. USA 106, 2313–2318 (2009).
Takeda, H., Takai, A., Inuzuka, T. & Marusawa, H. Genetic basis of hepatitis virus-associated hepatocellular carcinoma: linkage between infection, inflammation, and tumorigenesis. J. Gastroenterol. 52, 26–38 (2017).
McFadden, K. et al. Metabolic stress is a barrier to Epstein-Barr virus-mediated B cell immortalization. Proc. Natl Acad. Sci. USA 113, E782–790 (2016). This study discovers that metabolic and genotoxic stress experienced by EBV-infected cells represents a barrier to EBV-mediated transformation.
Nikitin, P. A. et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8, 510–522 (2010).
Li, J. et al. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 87, 9173–9188 (2013).
Shuda, M. et al. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl Acad. Sci. USA 105, 16272–16277 (2008).
Rosewick, N. et al. Cis-perturbation of cancer drivers by the HTLV-1/BLV proviruses is an early determinant of leukemogenesis. Nat. Commun. 8, 15264 (2017). In this study, HTLV-1 and bovine leukaemia virus proviruses are found to integrate preferentially near cancer driver genes and to cause their perturbation, favouring the expansion of the corresponding infected clone, allowing acquisition of additional somatic mutations that lead to the development of leukaemogenesis.
Nicot, C. HTLV-I Tax-mediated inactivation of cell cycle checkpoints and DNA repair pathways contribute to cellular transformation: “a random mutagenesis model”. J. Cancer Sci. 2, https://doi.org/10.13188/2377-9292.1000009 (2015).
Machida, K. et al. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc. Natl Acad. Sci. USA 101, 4262–4267 (2004). This study demonstrates that HCV infection induces a mutator phenotype to cause a significant increase in the mutation frequency of several proto-oncogenes, suggesting that HCV transforms cells by a hit-and-run mechanism.
Decorsiere, A. et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 531, 386–389 (2016).
Murphy, C. M. et al. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep. 16, 2846–2854 (2016). This study, together with reference 91, identifies the SMC complex SMC5/6 as a host restriction factor for HBV infection.
Thorley-Lawson, D. A. EBV persistence — introducing the virus. Curr. Top. Microbiol. Immunol. 390, 151–209 (2015).
Hopcraft, S. E. & Damania, B. Tumour viruses and innate immunity. Phil. Trans. R. Soc. B 372, 20160267 (2017).
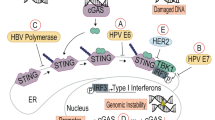
Chen, Q., Sun, L. & Chen, Z. J. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016).
Ma, Z. et al. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl Acad. Sci. USA 112, E4306–4315 (2015).
Wu, J. J. et al. Inhibition of cGAS DNA sensing by a herpesvirus virion protein. Cell Host Microbe 18, 333–344 (2015).
Zhang, G. et al. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl Acad. Sci. USA 113, E1034–E1043 (2016).
Guito, J. & Lukac, D. M. KSHV reactivation and novel implications of protein isomerization on lytic switch control. Viruses 7, 72–109 (2015).
Lau, L., Gray, E. E., Brunette, R. L. & Stetson, D. B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 350, 568–571 (2015). This study reveals that the oncogenes of the DNA tumour viruses, including HPV E7 and adenovirus E1A, can specifically inhibit the cGAS–STING pathway.
Yuen, C. K. et al. Suppression of type i interferon production by human T-cell leukemia virus type 1 oncoprotein Tax through Inhibition of IRF3 phosphorylation. J. Virol. 90, 3902–3912 (2016).
Liu, Y. et al. Hepatitis B virus polymerase disrupts K63-linked ubiquitination of STING to block innate cytosolic DNA-sensing pathways. J. Virol. 89, 2287–2300 (2015).
Nitta, S. et al. Hepatitis C virus NS4B protein targets STING and abrogates RIG-I-mediated type I interferon-dependent innate immunity. Hepatology 57, 46–58 (2013).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Yu, S. et al. Hepatitis B virus polymerase inhibits RIG-I− and Toll-like receptor 3-mediated β interferon induction in human hepatocytes through interference with interferon regulatory factor 3 activation and dampening of the interaction between TBK1/IKKε and DDX3. J. General Virol. 91, 2080–2090 (2010).
Wang, X. et al. Hepatitis B virus X protein suppresses virus-triggered IRF3 activation and IFN-β induction by disrupting the VISA-associated complex. Cell. Mol. Immunol. 7, 341–348 (2010).
Gregory, S. M. et al. Discovery of a viral NLR homolog that inhibits the inflammasome. Science 331, 330–334 (2011). In this study, KSHV ORF63 is discovered to be a viral homologue that inhibits human NLRP1-mediated innate immune response to promote KSHV reactivation and propagation.
Jacobs, S. R. & Damania, B. The viral interferon regulatory factors of KSHV: immunosuppressors or oncogenes? Frontiers Immunol. 2, 19 (2011).
Chatterjee, M., Osborne, J., Bestetti, G., Chang, Y. & Moore, P. S. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science 298, 1432–1435 (2002).
Taga, T. & Kishimoto, T. Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 15, 797–819 (1997).
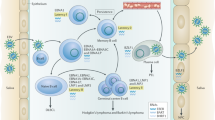
Hewitt, E. W. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology 110, 163–169 (2003).
Petersen, J. L., Morris, C. R. & Solheim, J. C. Virus evasion of MHC class I molecule presentation. J. Immunol. 171, 4473–4478 (2003).
Coscoy, L. & Ganem, D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl Acad. Sci. USA 97, 8051–8056 (2000).
Means, R. E., Ishido, S., Alvarez, X. & Jung, J. U. Multiple endocytic trafficking pathways of MHC class I molecules induced by a Herpesvirus protein. EMBO J. 21, 1638–1649 (2002).
Ishido, S., Wang, C., Lee, B. S., Cohen, G. B. & Jung, J. U. Downregulation of major histocompatibility complex class I molecules by Kaposi’s sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74, 5300–5309 (2000).
Banerjee, P., Feuer, G. & Barker, E. Human T cell leukemia virus type 1 (HTLV-1) p12I down-modulates ICAM-1 and -2 and reduces adherence of natural killer cells, thereby protecting HTLV-1-infected primary CD4+ T cells from autologous natural killer cell-mediated cytotoxicity despite the reduction of major histocompatibility complex class I molecules on infected cells. J. Virol. 81, 9707–9717 (2007).
Van Prooyen, N. et al. Human T cell leukemia virus type 1 p8 protein increases cellular conduits and virus transmission. Proc. Natl Acad. Sci. USA 107, 20738–20743 (2010).
Yasuma, K. et al. HTLV-1 bZIP factor impairs anti-viral immunity by inducing co-inhibitory molecule, T cell immunoglobulin and ITIM domain (TIGIT). PLoS Pathog. 12, e1005372 (2016).
Price, A. M. & Luftig, M. A. Dynamic Epstein-Barr virus gene expression on the path to B cell transformation. Adv. Virus Res. 88, 279–313 (2014).
Hatton, O. L., Harris-Arnold, A., Schaffert, S., Krams, S. M. & Martinez, O. M. The interplay between Epstein-Barr virus and B lymphocytes: implications for infection, immunity, and disease. Immunol. Res. 58, 268–276 (2014).
Caldwell, R. G., Wilson, J. B., Anderson, S. J. & Longnecker, R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9, 405–411 (1998).
Minamitani, T. et al. Evasion of affinity-based selection in germinal centers by Epstein-Barr virus LMP2A. Proc. Natl Acad. Sci. USA 112, 11612–11617 (2015).
Xia, T., Konno, H., Ahn, J. & Barber, G. N. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14, 282–297 (2016).
McQuaid, T., Savini, C. & Seyedkazemi, S. Sofosbuvir, a significant paradigm change in HCV treatment. J. Clin. Transl Hepatol. 3, 27–35 (2015).
Kaufman, H. L. et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet. Oncol. 17, 1374–1385 (2016).
Nghiem, P. T. et al. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N. Engl. J. Med. 374, 2542–2552 (2016).
Porta, C., Paglino, C. & Mosca, A. Targeting PI3K/Akt/mTOR signaling in cancer. Frontiers Oncol. 4, 64 (2014).
Dowling, R. J. et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176 (2010).
Chen, D., Jarrell, A., Guo, C., Lang, R. & Atit, R. Dermal β-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development 139, 1522–1533 (2012).
Eulalio, A., Huntzinger, E. & Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 132, 9–14 (2008).
Yates, L. A., Norbury, C. J. & Gilbert, R. J. The long and short of microRNA. Cell 153, 516–519 (2013).
Lujambio, A. & Lowe, S. W. The microcosmos of cancer. Nature 482, 347–355 (2012).
Gottwein, E. et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450, 1096–1099 (2007).
Gottwein, E. et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 10, 515–526 (2011).
Marquitz, A. R., Mathur, A., Shair, K. H. & Raab-Traub, N. Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc. Natl Acad. Sci. USA 109, 9593–9598 (2012).
Honegger, A. et al. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 11, e1004712 (2015).
Vernin, C. et al. HTLV-1 bZIP factor HBZ promotes cell proliferation and genetic instability by activating OncomiRs. Cancer Res. 74, 6082–6093 (2014).
Luna, J. M. et al. Hepatitis C virus RNA functionally sequesters miR-122. Cell 160, 1099–1110 (2015). This study suggests that HCV induced miR-122 sequestration, and subsequent derepression of host miR-122 targets promotes a cellular environment to support HCV-associated oncogenesis.
Raab-Traub, N. Novel mechanisms of EBV-induced oncogenesis. Curr. Opin. Virol. 2, 453–458 (2012).
Young, L. S. & Rickinson, A. B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4, 757–768 (2004).
Levrero, M. & Zucman-Rossi, J. Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol. 64, S84–101 (2016).
Gessain, A. & Cassar, O. Epidemiological aspects and world distribution of HTLV-1 infection. Front. Microbiol. 3, 388 (2012).
Matsuoka, M. & Jeang, K. T. Human T cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 7, 270–280 (2007).
Schiffman, M. et al. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers 2, 16086 (2016).
Harper, D. M. & DeMars, L. R. HPV vaccines — a review of the first decade. Gynecol. Oncol. 146, 196–204 (2017).
Mitchell, J. K., Lemon, S. M. & McGivern, D. R. How do persistent infections with hepatitis C virus cause liver cancer? Curr. Opin. Virol. 14, 101–108 (2015).
Goossens, N. & Hoshida, Y. Hepatitis C virus-induced hepatocellular carcinoma. Clin. Mol. Hepatol. 21, 105–114 (2015).
Schulz, T. F. & Cesarman, E. Kaposi Sarcoma-associated Herpesvirus: mechanisms of oncogenesis. Curr. Opin. Virol. 14, 116–128 (2015).
Wendzicki, J. A., Moore, P. S. & Chang, Y. Large T and small T antigens of Merkel cell polyomavirus. Curr. Opin. Virol. 11, 38–43 (2015).
Acknowledgements
The authors thank E. A. White, C. B. Buck and the members of the You laboratory for valuable comments and suggestions on the manuscript. The authors apologize to all colleagues whose primary research papers could not be cited owing to space constraints. Research on human papilloma virus (HPV) and Merkel cell polyomavirus (MCPyV) in the You laboratory has been supported by National Institutes of Health (NIH) grants (R01CA187718, R01CA148768 and R01CA142723) and the National Cancer Institute (NCI) Cancer Center Support grant (NCI P30 CA016520).
Reviewer information
Nature Reviews Microbiology thanks D. Galloway and other anonymous reviewers for their contributions to the peer review of this work.
Author information
Authors and Affiliations
Contributions
N.A.K. and J.Y. researched data for the article, substantially contributed to discussion of content, wrote the article and reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary terms
- Oncogenic viruses
-
Viruses that cause cancer. Sometimes also called tumour viruses. However, some tumour viruses, such as adenovirus and polyomavirus SV40 promote tumorigenesis in other organisms and infect humans but do not cause human cancers.
- Solid tumours
-
Masses of transformed and supporting cells that arise in stationary tissues (sarcomas and carcinomas) and not from cells of haematopoietic origin.
- Lymphoma
-
Tumour arising from a lymphoid cell type that occurs predominantly in the lymphatics, as opposed to leukaemias, in which the cancer cells are found in the blood.
- Cellular transformation
-
Selective acquisition of cellular traits, such as replicative immortality, increased stemness, growth factor independence, resistance to growth suppressors and alterations to metabolic flux.
- Metastasis
-
Tumour migration, invasion and colonization of body sites other than the primary site.
- p53
-
A transcription factor and key tumour suppressor downstream of exogenous signals and DNA damage-sensing pathways that maintains genome integrity and governs cell fate by promoting expression of effectors of DNA repair, cell cycle arrest, senescence and apoptosis.
- pRB
-
(Retinoblastoma protein). A tumour suppressor that is responsible for a major G1 checkpoint that blocks S-phase entry and cellular growth.
- Oncoproteins
-
Translated gene products that have the capacity to drive cellular transformation.
- Kaposi sarcoma
-
A family of endothelial malignancies that are associated with Kaposi sarcoma-associated virus (KSHV) and whose members are classified by the type of immunosuppression that enabled KSHV-mediated oncogenesis.
- Carcinomas
-
Tumours arising from cells of an epithelial origin, as opposed to sarcomas, which arise from mesenchymal cells.
- Rapamycin
-
An inhibitor of mechanistic target of rapamycin (mTOR)-mediated proliferative function that acts through direct binding of the peptidyl-prolyl cis-trans isomerase FKBP1A–mechanistic target of rapamycin complex and has shown promise as an immunosuppressant and antitumour drug.
- Sarcomagenesis
-
The seminal event or events leading to cancer progression from mesenchymal-derived cell types.
- Cap-dependent translation
-
Translation in which initiation is mediated by recognition of the 5′ cap that is specific to eukaryotic mRNAs.
- MEK1
-
(MAPK/ERK kinase 1 (also known as MAP2K1)). A crucial protein kinase that mediates an intermediate step of the RAF–MEK–ERK phosphorylation cascade responsible for activating expression of pro-proliferative, survival and differentiation genes in response to external stimuli.
- Invasive cells
-
Tumour cells with characteristics that enable them to metastasize and invade other tissues.
- Endothelial-to-mesenchymal transition
-
A process essential to cardiac development and normal angiogenesis by which endothelial cells acquire stem-like, mesenchymal traits including enhanced migration that, in certain cancers, contribute to metastatic ability.
- Proto-oncogene
-
A type of endogenous gene that, when overexpressed or abnormally activated as a result of mutation, can promote cancer development, at which point it is referred to as an oncogene.
- CHK2
-
(Checkpoint kinase 2). A tumour suppressor kinase activated by ataxia telangiectasia mutated (ATM) in response to double-stranded breaks in DNA that maintains genomic integrity by mediating cell cycle arrest and DNA repair.
- Second messengers
-
Soluble small molecules that transduce intracellular signals, which can be secreted by intracellular bacteria to coordinate responses to their environment.
- Inflammasome
-
A cytoplasmic complex of NOD-, LRR- and pyrin domain-containing proteins (NLRPs), adaptor proteins and caspases that forms in response to cellular damage or bacterial effectors that cause rapid caspase-mediated inflammatory cytokine release and/or a type of lytic cell death called pyroptosis.
- T cell anergy
-
A process by which CD4+ or CD8+ T cells become tolerant to antigens and functionally inactivated owing to stimulation in the absence of a necessary signal.
- CHK1
-
(Checkpoint kinase 1). A protein kinase essential to normal cell division and development that is activated by ataxia telangiectasia and Rad3-related protein (ATR) in response to single-stranded DNA to facilitate proper DNA replication, cell cycle progression and response to DNA insults.
- Seed sequence
-
An eight-nucleotide sequence near the 5′-end of a microRNA that undergoes Watson–Crick base pairing with a target RNA with high specificity and that is required for efficient targeting.
Rights and permissions
About this article
Cite this article
Krump, N.A., You, J. Molecular mechanisms of viral oncogenesis in humans. Nat Rev Microbiol 16, 684–698 (2018). https://doi.org/10.1038/s41579-018-0064-6
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-018-0064-6
This article is cited by
-
Navigating therapeutic strategies: HPV classification in head and neck cancer
British Journal of Cancer (2024)
-
Intratumoural microbiota: a new frontier in cancer development and therapy
Signal Transduction and Targeted Therapy (2024)
-
Viruses in glioblastoma: an update on evidence and clinical trials
BJC Reports (2024)
-
Abnormal protein SUMOylation in liver disease: novel target for therapy
Journal of Molecular Medicine (2024)
-
The Microbiome Matters: Its Impact on Cancer Development and Therapeutic Responses
Journal of Microbiology (2024)