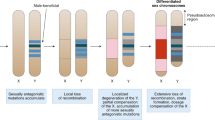
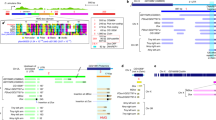
Abstract
Sexually reproducing eukaryotes use recombination between homologous chromosomes to promote chromosome segregation during meiosis. Meiotic recombination is almost universally conserved in its broad strokes, but specific molecular details often differ considerably between taxa, and the proteins that constitute the recombination machinery show substantial sequence variability. The extent of this variation is becoming increasingly clear because of recent increases in genomic resources and advances in protein structure prediction. We discuss the tension between functional conservation and rapid evolutionary change with a focus on the proteins that are required for the formation and repair of meiotic DNA double-strand breaks. We highlight phylogenetic relationships on different time scales and propose that this remarkable evolutionary plasticity is a fundamental property of meiotic recombination that shapes our understanding of molecular mechanisms in reproductive biology.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
de Massy, B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47, 563–599 (2013).
Keeney, S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53 (2001).
Hunter, N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015).
Hassold, T. J. & Hunt, P. A. Missed connections: recombination and human aneuploidy. Prenat. Diagn. 41, 584–590 (2021).
Boekhout, M. et al. REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell 74, 1053–1068.e1058 (2019).
Dapper, A. L. & Payseur, B. A. Molecular evolution of the meiotic recombination pathway in mammals. Evolution 73, 2368–2389 (2019). This study uses molecular evolution analyses to systematically characterize components of the mammalian recombination machinery and finds evidence for rapid evolution in key components.
Keeney, S. Spo11 and the formation of DNA double-strand breaks in meiosis. Genome Dyn. Stab. 2, 81–123 (2008).
Murat, F. et al. The molecular evolution of spermatogenesis across mammals. Nature 613, 308–316 (2023). Using single-nucleus transcriptomics across different mammalian species, this study characterizes changes in spermatogenesis at unprecedented resolution.
Malik, S. B., Ramesh, M. A., Hulstrand, A. M. & Logsdon, J. M. Jr. Protist homologs of the meiotic Spo11 gene and topoisomerase VI reveal an evolutionary history of gene duplication and lineage-specific loss. Mol. Biol. Evol. 24, 2827–2841 (2007). Using degenerate PCR and database searches this study identifies homologues of SPO11 and TOP6BL in protists.
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997).
Bergerat, A. et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417 (1997).
Robert, T. et al. The TOPOVIB-like protein family is required for meiotic DNA double-strand break formation. Science 351, 943–949 (2016). This study identified the SPO11 partner TOPOVIBL in mouse.
Vrielynck, N. et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351, 939–943 (2016). This study identified the SPO11 partner TOPOVIBL in Arabidopsis.
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Fraune, J., Wiesner, M. & Benavente, R. The synaptonemal complex of basal metazoan hydra: more similarities to vertebrate than invertebrate meiosis model organisms. J. Genet. Genomics 41, 107–115 (2014).
Loidl, J. Conservation and variability of meiosis across the eukaryotes. Annu. Rev. Genet. 50, 293–316 (2016).
Ishiguro, K. I. The cohesin complex in mammalian meiosis. Genes Cells 24, 6–30 (2019).
Zickler, D. & Kleckner, N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb. Perspect. Biol. 7, a016626 (2015).
Longhese, M. P., Bonetti, D., Guerini, I., Manfrini, N. & Clerici, M. DNA double-strand breaks in meiosis: checking their formation, processing and repair. DNA Repair 8, 1127–1138 (2009).
Gerton, J. L. & Hawley, R. S. Homologous chromosome interactions in meiosis: diversity amidst conservation. Nat. Rev. Genet. 6, 477–487 (2005).
de Boer, E. & Heyting, C. The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115, 220–234 (2006).
Subramanian, V. V. & Hochwagen, A. The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 6, a016675 (2014).
Ivanov, E. L., Korolev, V. G. & Fabre, F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132, 651–664 (1992).
Engebrecht, J., Hirsch, J. & Roeder, G. S. Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell 62, 927–937 (1990).
Malone, R. E. et al. Isolation of mutants defective in early steps of meiotic recombination in the yeast Saccharomyces cerevisiae. Genetics 128, 79–88 (1991).
Menees, T. M. & Roeder, G. S. MEI4, a yeast gene required for meiotic recombination. Genetics 123, 675–682 (1989).
Gardiner, J. M., Bullard, S. A., Chrome, C. & Malone, R. E. Molecular and genetic analysis of REC103, an early meiotic recombination gene in yeast. Genetics 146, 1265–1274 (1997).
Bhargava, J., Engebrecht, J. & Roeder, G. S. The rec102 mutant of yeast is defective in meiotic recombination and chromosome synapsis. Genetics 130, 59–69 (1992).
Galbraith, A. M. & Malone, R. E. Characterization of REC104, a gene required for early meiotic recombination in the yeast Saccharomyces cerevisiae. Dev. Genet. 13, 392–402 (1992).
Johzuka, K. & Ogawa, H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics 139, 1521–1532 (1995).
Game, J. C., Zamb, T. J., Braun, R. J., Resnick, M. & Roth, R. M. The role of radiation (rad) genes in meiotic recombination in yeast. Genetics 94, 51–68 (1980).
Jiao, K., Salem, L. & Malone, R. Support for a meiotic recombination initiation complex: interactions among Rec102p, Rec104p, and Spo11p. Mol. Cell Biol. 23, 5928–5938 (2003).
Kee, K. & Keeney, S. Functional interactions between SPO11 and REC102 during initiation of meiotic recombination in Saccharomyces cerevisiae. Genetics 160, 111–122 (2002).
Kee, K., Protacio, R. U., Arora, C. & Keeney, S. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 23, 1815–1824 (2004).
Claeys Bouuaert, C. et al. Structural and functional characterization of the Spo11 core complex. Nat. Struct. Mol. Biol. 28, 92–102 (2021).
Arora, C., Kee, K., Maleki, S. & Keeney, S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13, 549–559 (2004).
Claeys Bouuaert, C. et al. DNA-driven condensation assembles the meiotic DNA break machinery. Nature 592, 144–149 (2021).
Neale, M. J., Pan, J. & Keeney, S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057 (2005).
Garcia, V., Phelps, S. E., Gray, S. & Neale, M. J. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479, 241–244 (2011).
Prinz, S., Amon, A. & Klein, F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics 146, 781–795 (1997).
McKee, A. H. & Kleckner, N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics 146, 797–816 (1997).
Chin, G. M. & Villeneuve, A. M. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 15, 522–534 (2001).
Goodyer, W. et al. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev. Cell 14, 263–274 (2008).
Yin, Y. & Smolikove, S. Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol. Cell Biol. 33, 2732–2747 (2013).
Bishop, D. K., Park, D., Xu, L. & Kleckner, N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell 69, 439–456 (1992).
Shinohara, A., Ogawa, H. & Ogawa, T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell 69, 457–470 (1992).
Lin, Z., Kong, H., Nei, M. & Ma, H. Origins and evolution of the recA/RAD51 gene family: evidence for ancient gene duplication and endosymbiotic gene transfer. Proc. Natl Acad. Sci. USA 103, 10328–10333 (2006). An evolutionary characterization of the RAD51 gene family.
Tang, S., Wu, M. K. Y., Zhang, R. & Hunter, N. Pervasive and essential roles of the Top3-Rmi1 decatenase orchestrate recombination and facilitate chromosome segregation in meiosis. Mol. Cell 57, 607–621 (2015).
Kaur, H., De Muyt, A. & Lichten, M. Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates. Mol. Cell 57, 583–594 (2015).
De Muyt, A. et al. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46, 43–53 (2012).
Amin, A. D., Chaix, A. B., Mason, R. P., Badge, R. M. & Borts, R. H. The roles of the Saccharomyces cerevisiae RecQ helicase SGS1 in meiotic genome surveillance. PLoS ONE 5, e15380 (2010).
Oh, S. D. et al. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130, 259–272 (2007).
Fasching, C. L., Cejka, P., Kowalczykowski, S. C. & Heyer, W. D. Top3-Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol. Cell 57, 595–606 (2015).
Sandhu, R. et al. DNA helicase Mph1(FANCM) ensures meiotic recombination between parental chromosomes by dissociating precocious displacement loops. Dev. Cell 53, 458–472 (2020).
Sasanuma, H. et al. Srs2 helicase prevents the formation of toxic DNA damage during late prophase I of yeast meiosis. Chromosoma 128, 453–471 (2019).
Palladino, F. & Klein, H. L. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132, 23–37 (1992).
Zakharyevich, K., Tang, S., Ma, Y. & Hunter, N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149, 334–347 (2012).
Zakharyevich, K. et al. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol. Cell 40, 1001–1015 (2010).
Cannavo, E. et al. Regulation of the MLH1-MLH3 endonuclease in meiosis. Nature 586, 618–622 (2020).
Nishant, K. T., Plys, A. J. & Alani, E. A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae. Genetics 179, 747–755 (2008).
Argueso, J. L. et al. Analysis of conditional mutations in the Saccharomyces cerevisiae MLH1 gene in mismatch repair and in meiotic crossing over. Genetics 160, 909–921 (2002).
Rockmill, B., Fung, J. C., Branda, S. S. & Roeder, G. S. The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13, 1954–1962 (2003).
Ross-Macdonald, P. & Roeder, G. S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79, 1069–1080 (1994).
Hollingsworth, N. M., Ponte, L. & Halsey, C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9, 1728–1739 (1995).
Mazina, O. M., Mazin, A. V., Nakagawa, T., Kolodner, R. D. & Kowalczykowski, S. C. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117, 47–56 (2004).
De Muyt, A. et al. A meiotic XPF-ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 32, 283–296 (2018).
Chua, P. R. & Roeder, G. S. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93, 349–359 (1998).
Börner, G. V., Kleckner, N. & Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45 (2004).
Agarwal, S. & Roeder, G. S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102, 245–255 (2000).
Berchowitz, L. E. & Copenhaver, G. P. Genetic interference: don’t stand so close to me. Curr. Genomics 11, 91–102 (2010).
Argueso, J. L., Wanat, J., Gemici, Z. & Alani, E. Competing crossover pathways act during meiosis in Saccharomyces cerevisiae. Genetics 168, 1805–1816 (2004).
de los Santos, T. et al. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164, 81–94 (2003).
de los Santos, T., Loidl, J., Larkin, B. & Hollingsworth, N. M. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics 159, 1511–1525 (2001).
Lin, Y. & Smith, G. R. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics 136, 769–779 (1994).
Cervantes, M. D., Farah, J. A. & Smith, G. R. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5, 883–888 (2000).
Miyoshi, T. et al. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell 47, 722–733 (2012).
Evans, D. H., Li, Y. F., Fox, M. E. & Smith, G. R. A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics 146, 1253–1264 (1997).
Lin, Y. & Smith, G. R. An intron-containing meiosis-induced recombination gene, rec15, of Schizosaccharomyces pombe. Mol. Microbiol. 17, 439–448 (1995).
Bonfils, S., Rozalen, A. E., Smith, G. R., Moreno, S. & Martin-Castellanos, C. Functional interactions of Rec24, the fission yeast ortholog of mouse Mei4, with the meiotic recombination-initiation complex. J. Cell Sci. 124, 1328–1338 (2011).
Molnar, M. et al. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157, 519–532 (2001).
Steiner, S., Kohli, J. & Ludin, K. Functional interactions among members of the meiotic initiation complex in fission yeast. Curr. Genet. 56, 237–249 (2010).
Young, J. A., Hyppa, R. W. & Smith, G. R. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167, 593–605 (2004).
Gregan, J. et al. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 15, 1663–1669 (2005).
Thompson, E. A. & Roeder, G. S. Expression and DNA sequence of RED1, a gene required for meiosis I chromosome segregation in yeast. Mol. Gen. Genet. 218, 293–301 (1989).
Rockmill, B. & Roeder, G. S. Meiosis in asynaptic yeast. Genetics 126, 563–574 (1990).
Tavassoli, M., Shayeghi, M., Nasim, A. & Watts, F. Z. Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23, 383–388 (1995).
Davidson, M. K., Young, N. P., Glick, G. G. & Wahls, W. P. Meiotic chromosome segregation mutants identified by insertional mutagenesis of fission yeast Schizosaccharomyces pombe; tandem-repeat, single-site integrations. Nucleic Acids Res. 32, 4400–4410 (2004).
Hartsuiker, E. et al. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell Biol. 29, 1671–1681 (2009).
Milman, N., Higuchi, E. & Smith, G. R. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell Biol. 29, 5998–6005 (2009).
Rothenberg, M., Kohli, J. & Ludin, K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 5, e1000722 (2009).
Farah, J. A., Cromie, G. A. & Smith, G. R. Ctp1 and exonuclease 1, alternative nucleases regulated by the MRN complex, are required for efficient meiotic recombination. Proc. Natl Acad. Sci. USA 106, 9356–9361 (2009).
Grishchuk, A. L. & Kohli, J. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics 165, 1031–1043 (2003).
Lorenz, A. et al. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336, 1585–1588 (2012).
Cromie, G. A., Hyppa, R. W. & Smith, G. R. The fission yeast BLM homolog Rqh1 promotes meiotic recombination. Genetics 179, 1157–1167 (2008).
Hope, J. C. et al. Mus81-Eme1-dependent and -independent crossovers form in mitotic cells during double-strand break repair in Schizosaccharomyces pombe. Mol. Cell Biol. 27, 3828–3838 (2007).
Boddy, M. N. et al. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548 (2001).
Cromie, G. A. et al. Single Holliday junctions are intermediates of meiotic recombination. Cell 127, 1167–1178 (2006).
McKim, K. S. & Hayashi-Hagihara, A. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 12, 2932–2942 (1998).
Liu, H., Jang, J. K., Kato, N. & McKim, K. S. mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162, 245–258 (2002).
Lake, C. M. et al. Vilya, a component of the recombination nodule, is required for meiotic double-strand break formation in Drosophila. eLife 4, e08287 (2015).
Lake, C. M., Nielsen, R. J. & Hawley, R. S. The Drosophila zinc finger protein trade embargo is required for double strand break formation in meiosis. PLoS Genet. 7, e1002005 (2011).
Lake, C. M. et al. Narya, a RING finger domain-containing protein, is required for meiotic DNA double-strand break formation and crossover maturation in Drosophila melanogaster. PLoS Genet. 15, e1007886 (2019).
Digilio, F. A., Pannuti, A., Lucchesi, J. C., Furia, M. & Polito, L. C. Tosca: a Drosophila gene encoding a nuclease specifically expressed in the female germline. Dev. Biol. 178, 90–100 (1996).
Yoo, S. & McKee, B. D. Functional analysis of the Drosophila Rad51 gene (spn-A) in repair of DNA damage and meiotic chromosome segregation. DNA Repair 4, 231–242 (2005).
Blanton, H. & Sekelsky, J. Unique invasions and resolutions: DNA repair proteins in meiotic recombination in Drosophila melanogaster. Cytogenet. Genome Res. 107, 172–179 (2004).
McVey, M., Andersen, S. L., Broze, Y. & Sekelsky, J. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176, 1979–1992 (2007).
Carpenter, A. T. & Sandler, L. On recombination-defective meiotic mutants in Drosophila melanogaster. Genetics 76, 453–475 (1974).
Grell, R. F. Time of recombination in the Drosophila melanogaster oocyte. III. selection and characterization of temperature-sensitive and -insensitive, recombination-deficient alleles in Drosophila. Genetics 108, 425–443 (1984).
Baker, B. S. & Carpenter, A. T. Genetic analysis of sex chromosomal meiotic mutants in Drosophilia melanogaster. Genetics 71, 255–286 (1972).
Blanton, H. L. et al. REC, Drosophila MCM8, drives formation of meiotic crossovers. PLoS Genet. 1, e40 (2005).
Page, S. L. et al. A germline clone screen for meiotic mutants in Drosophila melanogaster. Fly 1, 172–181 (2007).
Lake, C. M., Teeter, K., Page, S. L., Nielsen, R. & Hawley, R. S. A genetic analysis of the Drosophila mcm5 gene defines a domain specifically required for meiotic recombination. Genetics 176, 2151–2163 (2007).
Kohl, K. P., Jones, C. D. & Sekelsky, J. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338, 1363–1365 (2012).
Green, M. M. mus(3)312D1, a mutagen sensitive mutant with profound effects on female meiosis in Drosophila melanogaster. Chromosoma 82, 259–266 (1981).
Radford, S. J., Goley, E., Baxter, K., McMahan, S. & Sekelsky, J. Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170, 1737–1745 (2005).
Joyce, E. F., Tanneti, S. N. & McKim, K. S. Drosophila hold’em is required for a subset of meiotic crossovers and interacts with the dna repair endonuclease complex subunits MEI-9 and ERCC1. Genetics 181, 335–340 (2009).
Trowbridge, K., McKim, K., Brill, S. J. & Sekelsky, J. Synthetic lethality of Drosophila in the absence of the MUS81 endonuclease and the DmBlm helicase is associated with elevated apoptosis. Genetics 176, 1993–2001 (2007).
Dernburg, A. F. et al. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387–398 (1998).
Hinman, A. W. et al. Caenorhabditis elegans DSB-3 reveals conservation and divergence among protein complexes promoting meiotic double-strand breaks. Proc. Natl Acad. Sci. USA 118, e2109306118 (2021).
Stamper, E. L. et al. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9, e1003679 (2013).
Rosu, S. et al. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 9, e1003674 (2013).
Zetka, M. C., Kawasaki, I., Strome, S. & Muller, F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13, 2258–2270 (1999).
Kim, Y. et al. The chromosome axis controls meiotic events through a hierarchical assembly of HORMA domain proteins. Dev. Cell 31, 487–502 (2014).
Couteau, F. & Zetka, M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 19, 2744–2756 (2005).
Fukuda, T., Daniel, K., Wojtasz, L., Toth, A. & Hoog, C. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp. Cell Res. 316, 158–171 (2010).
Shin, Y. H. et al. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 6, e1001190 (2010).
Hollingsworth, N. M. & Byers, B. HOP1: a yeast meiotic pairing gene. Genetics 121, 445–462 (1989).
Caryl, A. P., Armstrong, S. J., Jones, G. H. & Franklin, F. C. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109, 62–71 (2000).
Latypov, V. et al. Roles of Hop1 and Mek1 in meiotic chromosome pairing and recombination partner choice in Schizosaccharomyces pombe. Mol. Cell Biol. 30, 1570–1581 (2010).
Hayashi, M., Chin, G. M. & Villeneuve, A. M. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3, e191 (2007).
Girard, C., Roelens, B., Zawadzki, K. A. & Villeneuve, A. M. Interdependent and separable functions of Caenorhabditis elegans MRN-C complex members couple formation and repair of meiotic DSBs. Proc. Natl Acad. Sci. USA 115, E4443–E4452 (2018).
Penkner, A. et al. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 26, 5071–5082 (2007).
Alpi, A., Pasierbek, P., Gartner, A. & Loidl, J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112, 6–16 (2003).
Rinaldo, C., Bazzicalupo, P., Ederle, S., Hilliard, M. & La Volpe, A. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160, 471–479 (2002).
Youds, J. L. et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327, 1254–1258 (2010).
Zhang, L., Kohler, S., Rillo-Bohn, R. & Dernburg, A. F. A compartmentalized signaling network mediates crossover control in meiosis. eLife 7, e30789 (2018).
Nguyen, H., Labella, S., Silva, N., Jantsch, V. & Zetka, M. C. elegans ZHP-4 is required at multiple distinct steps in the formation of crossovers and their transition to segregation competent chiasmata. PLoS Genet. 14, e1007776 (2018).
Zalevsky, J., MacQueen, A. J., Duffy, J. B., Kemphues, K. J. & Villeneuve, A. M. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153, 1271–1283 (1999).
Yokoo, R. et al. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012).
Agostinho, A. et al. Combinatorial regulation of meiotic holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9, e1003591 (2013).
O’Neil, N. J. et al. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 9, e1003582 (2013).
Saito, T. T., Lui, D. Y., Kim, H. M., Meyer, K. & Colaiacovo, M. P. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9, e1003586 (2013).
Wicky, C. et al. Multiple genetic pathways involving the Caenorhabditis elegans Bloom’s syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Mol. Cell Biol. 24, 5016–5027 (2004).
Romanienko, P. J. & Camerini-Otero, R. D. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell 6, 975–987 (2000).
Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6, 989–998 (2000).
Stanzione, M. et al. Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 18, 1208–1220 (2016).
Kumar, R., Bourbon, H. M. & de Massy, B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24, 1266–1280 (2010).
Libby, B. J., Reinholdt, L. G. & Schimenti, J. C. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc. Natl Acad. Sci. USA 100, 15706–15711 (2003).
Cherry, S. M. et al. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr. Biol. 17, 373–378 (2007).
Zhang, B., Tang, Z., Li, L. & Lu, L. Y. NBS1 is required for SPO11-linked DNA double-strand break repair in male meiosis. Cell Death Differ. 27, 2176–2190 (2020).
Yamada, S. et al. Molecular structures and mechanisms of DNA break processing in mouse meiosis. Genes Dev. 34, 806–818 (2020).
Paiano, J. et al. ATM and PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nat. Commun. 11, 857 (2020).
Dai, J., Voloshin, O., Potapova, S. & Camerini-Otero, R. D. Meiotic knockdown and complementation reveals essential role of RAD51 in mouse spermatogenesis. Cell Rep. 18, 1383–1394 (2017).
Pittman, D. L. et al. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell 1, 697–705 (1998).
Yoshida, K. et al. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell 1, 707–718 (1998).
La Salle, S. et al. Spata22, a novel vertebrate-specific gene, is required for meiotic progress in mouse germ cells. Biol. Reprod. 86, 45 (2012).
Luo, M. et al. MEIOB exhibits single-stranded DNA-binding and exonuclease activities and is essential for meiotic recombination. Nat. Commun. 4, 2788 (2013).
Souquet, B. et al. MEIOB targets single-strand DNA and is necessary for meiotic recombination. PLoS Genet. 9, e1003784 (2013).
Holloway, J. K., Morelli, M. A., Borst, P. L. & Cohen, P. E. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J. Cell Biol. 188, 779–789 (2010).
de Vries, S. S. et al. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 13, 523–531 (1999).
Kneitz, B. et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 14, 1085–1097 (2000).
Guiraldelli, M. F., Eyster, C., Wilkerson, J. L., Dresser, M. E. & Pezza, R. J. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet. 9, e1003383 (2013).
Lipkin, S. M. et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat. Genet. 31, 385–390 (2002).
Edelmann, W. et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21, 123–127 (1999).
Baker, S. M. et al. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13, 336–342 (1996).
Edelmann, W. et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell 85, 1125–1134 (1996).
Kadri, N. K. et al. Coding and noncoding variants in HFM1, MLH3, MSH4, MSH5, RNF212, and RNF212B affect recombination rate in cattle. Genome Res. 26, 1323–1332 (2016).
Johnston, S. E., Huisman, J. & Pemberton, J. M. A genomic region containing REC8 and RNF212B is associated with individual recombination rate variation in a wild population of red deer (Cervus elaphus). G3 8, 2265–2276 (2018).
Johnston, S. E., Berenos, C., Slate, J. & Pemberton, J. M. Conserved genetic architecture underlying individual recombination rate variation in a wild population of soay sheep (Ovis aries). Genetics 203, 583–598 (2016).
Holloway, J. K., Sun, X., Yokoo, R., Villeneuve, A. M. & Cohen, P. E. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J. Cell Biol. 205, 633–641 (2014).
Guiraldelli, M. F. et al. SHOC1 is a ERCC4-(HhH)2-like protein, integral to the formation of crossover recombination intermediates during mammalian meiosis. PLoS Genet. 14, e1007381 (2018).
Reynolds, A. et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat. Genet. 45, 269–278 (2013).
Adelman, C. A. & Petrini, J. H. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 4, e1000042 (2008).
Yang, F. et al. Meiotic failure in male mice lacking an X-linked factor. Genes Dev. 22, 682–691 (2008).
Ward, J. O. et al. Mutation in mouse Hei10, an E3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 3, 1550–1563 (2007).
Zhang, Q., Ji, S. Y., Busayavalasa, K. & Yu, C. SPO16 binds SHOC1 to promote homologous recombination and crossing-over in meiotic prophase I. Sci. Adv. 5, eaau9780 (2019).
Holloway, J. K., Booth, J., Edelmann, W., McGowan, C. H. & Cohen, P. E. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4, e1000186 (2008).
Grelon, M., Vezon, D., Gendrot, G. & Pelletier, G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 20, 589–600 (2001).
Stacey, N. J. et al. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant. J. 48, 206–216 (2006).
Hartung, F. & Puchta, H. Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res. 28, 1548–1554 (2000).
Sugimoto-Shirasu, K., Stacey, N. J., Corsar, J., Roberts, K. & McCann, M. C. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr. Biol. 12, 1782–1786 (2002).
Hartung, F. et al. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr. Biol. 12, 1787–1791 (2002).
De Muyt, A. et al. AtPRD1 is required for meiotic double strand break formation in Arabidopsis thaliana. EMBO J. 26, 4126–4137 (2007).
De Muyt, A. et al. A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 5, e1000654 (2009).
Zhang, C. et al. The Arabidopsis thaliana DSB formation (AtDFO) gene is required for meiotic double-strand break formation. Plant. J. 72, 271–281 (2012).
Jolivet, S., Vezon, D., Froger, N. & Mercier, R. Non conservation of the meiotic function of the Ski8/Rec103 homolog in Arabidopsis. Genes Cells 11, 615–622 (2006). This study finds that the Ski8 homologue in A. thaliana Rec103 is not required for meiotic DSB formation, highlighting the importance of rigourous functional studies of homologues across model organisms.
Puizina, J., Siroky, J., Mokros, P., Schweizer, D. & Riha, K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant. Cell 16, 1968–1978 (2004).
Gallego, M. E. et al. Disruption of the Arabidopsis RAD50 gene leads to plant sterility and MMS sensitivity. Plant. J. 25, 31–41 (2001).
Bleuyard, J. Y., Gallego, M. E. & White, C. I. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113, 197–203 (2004).
Waterworth, W. M. et al. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant. J. 52, 41–52 (2007).
Uanschou, C. et al. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 26, 5061–5070 (2007).
Couteau, F. et al. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant. Cell 11, 1623–1634 (1999).
Li, W. et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl Acad. Sci. USA 101, 10596–10601 (2004).
Hartung, F., Suer, S. & Puchta, H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 104, 18836–18841 (2007).
Higgins, J. D., Ferdous, M., Osman, K. & Franklin, F. C. The RecQ helicase AtRECQ4A is required to remove inter-chromosomal telomeric connections that arise during meiotic recombination in Arabidopsis. Plant. J. 65, 492–502 (2011).
Hartung, F., Suer, S., Knoll, A., Wurz-Wildersinn, R. & Puchta, H. Topoisomerase 3α and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet. 4, e1000285 (2008).
Chelysheva, L., Vezon, D., Belcram, K., Gendrot, G. & Grelon, M. The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet. 4, e1000309 (2008).
Crismani, W. et al. FANCM limits meiotic crossovers. Science 336, 1588–1590 (2012).
Chelysheva, L. et al. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 3, e83 (2007).
Macaisne, N. et al. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Curr. Biol. 18, 1432–1437 (2008).
Macaisne, N., Vignard, J. & Mercier, R. SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J. Cell Sci. 124, 2687–2691 (2011).
Lu, P., Wijeratne, A. J., Wang, Z., Copenhaver, G. P. & Ma, H. Arabidopsis PTD is required for type I crossover formation and affects recombination frequency in two different chromosomal regions. J. Genet. Genomics 41, 165–175 (2014).
Wijeratne, A. J., Chen, C., Zhang, W., Timofejeva, L. & Ma, H. The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol. Biol. Cell 17, 1331–1343 (2006).
Higgins, J. D. et al. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. Plant. J. 55, 28–39 (2008).
Lu, X. et al. The Arabidopsis MutS homolog AtMSH5 is required for normal meiosis. Cell Res. 18, 589–599 (2008).
Higgins, J. D., Armstrong, S. J., Franklin, F. C. & Jones, G. H. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 18, 2557–2570 (2004).
Chen, C., Zhang, W., Timofejeva, L., Gerardin, Y. & Ma, H. The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant. J. 43, 321–334 (2005).
Mercier, R. et al. Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3,whereas the other one is not. Curr. Biol. 15, 692–701 (2005).
Chelysheva, L. et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012).
Dion, E., Li, L., Jean, M. & Belzile, F. An Arabidopsis MLH1 mutant exhibits reproductive defects and reveals a dual role for this gene in mitotic recombination. Plant. J. 51, 431–440 (2007).
Jackson, N. et al. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 25, 1315–1323 (2006).
Berchowitz, L. E., Francis, K. E., Bey, A. L. & Copenhaver, G. P. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3, e132 (2007).
Thangavel, G., Hofstatter, P. G., Mercier, R. & Marques, A. Tracing the evolution of the plant meiotic molecular machinery. Plant Reprod. 36, 73–95 (2023).
Tzung, K. W. et al. Genomic evidence for a complete sexual cycle in Candida albicans. Proc. Natl Acad. Sci. USA 98, 3249–3253 (2001).
Bloomfield, G., Paschke, P., Okamoto, M., Stevens, T. J. & Urushihara, H. Triparental inheritance in Dictyostelium. Proc. Natl Acad. Sci. USA 116, 2187–2192 (2019).
Forterre, P. & Gadelle, D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 37, 679–692 (2009).
Corbett, K. D., Benedetti, P. & Berger, J. M. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat. Struct. Mol. Biol. 14, 611–619 (2007).
Brinkmeier, J., Coelho, S., de Massy, B. & Bourbon, H. M. Evolution and diversity of the TopoVI and TopoVI-like subunits with extensive divergence of the TOPOVIBL subunit. Mol. Biol. Evol. 39, msac227 (2022). This study investigates the phylogenetic conservation of TopoVI and TopoVI-like subunits.
Ronceret, A., Doutriaux, M. P., Golubovskaya, I. N. & Pawlowski, W. P. PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc. Natl Acad. Sci. USA 106, 20121–20126 (2009).
Vrielynck, N. et al. Conservation and divergence of meiotic DNA double strand break forming mechanisms in Arabidopsis thaliana. Nucleic Acids Res. 49, 9821–9835 (2021). This study investigates the roles of different components of the meiotic DSB machinery in A. thaliana with a focus on evolutionary conservation and divergence.
Pawlowski, W. P. et al. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science 303, 89–92 (2004).
Tesse, S., Storlazzi, A., Kleckner, N., Gargano, S. & Zickler, D. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl Acad. Sci. USA 100, 12865–12870 (2003).
Johnson, A. W. & Kolodner, R. D. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol. Cell Biol. 15, 2719–2727 (1995).
Yuan, L. et al. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell 5, 73–83 (2000).
Ferdous, M. et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 8, e1002507 (2012).
Lemmens, B. B., Johnson, N. M. & Tijsterman, M. COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet. 9, e1003276 (2013).
Ribeiro, J., Abby, E., Livera, G. & Martini, E. RPA homologs and ssDNA processing during meiotic recombination. Chromosoma 125, 265–276 (2016).
Cloud, V., Chan, Y. L., Grubb, J., Budke, B. & Bishop, D. K. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science 337, 1222–1225 (2012).
Da Ines, O. et al. Meiotic recombination in Arabidopsis is catalysed by DMC1, with RAD51 playing a supporting role. PLoS Genet. 9, e1003787 (2013).
Wang, S. et al. Role of EXO1 nuclease activity in genome maintenance, the immune response and tumor suppression in Exo1D173A mice. Nucleic Acids Res. 50, 8093–8106 (2022).
Kulkarni, D. S. et al. PCNA activates the MutLγ endonuclease to promote meiotic crossing over. Nature 586, 623–627 (2020).
Gioia, M. et al. Exo1 protects DNA nicks from ligation to promote crossover formation during meiosis. PLoS Biol. 21, e3002085 (2023).
Shannon, M. et al. Characterization of the mouse Xpf DNA repair gene and differential expression during spermatogenesis. Genomics 62, 427–435 (1999).
Kirkpatrick, D. T. Roles of the DNA mismatch repair and nucleotide excision repair proteins during meiosis. Cell Mol. Life Sci. 55, 437–449 (1999).
Mastro, T. L. & Forsburg, S. L. Increased meiotic crossovers and reduced genome stability in absence of Schizosaccharomyces pombe Rad16 (XPF). Genetics 198, 1457–1472 (2014).
Hsia, K. T. et al. DNA repair gene Ercc1 is essential for normal spermatogenesis and oogenesis and for functional integrity of germ cell DNA in the mouse. Development 130, 369–378 (2003).
Meneely, P. M., Farago, A. F. & Kauffman, T. M. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics 162, 1169–1177 (2002).
Weinstein, A. The geometry and mechanics of crossing over. Cold Spring Harb. Symp. Quant. Biol. 23, 177–196 (1958).
Crismani, W. et al. MCM8 is required for a pathway of meiotic double-strand break repair independent of DMC1 in Arabidopsis thaliana. PLoS Genet. 9, e1003165 (2013).
Rao, H. B. et al. A SUMO-ubiquitin relay recruits proteasomes to chromosome axes to regulate meiotic recombination. Science 355, 403–407 (2017).
Swanson, W. J. & Vacquier, V. D. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 (2002).
Clark, N. L., Aagaard, J. E. & Swanson, W. J. Evolution of reproductive proteins from animals and plants. Reproduction 131, 11–22 (2006).
Civetta, A. & Singh, R. S. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in Drosophila melanogaster and Drosophila virilis group species. J. Mol. Evol. 41, 1085–1095 (1995).
Haerty, W. et al. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics 177, 1321–1335 (2007).
Coulthart, M. B. & Singh, R. S. High level of divergence of male-reproductive-tract proteins, between Drosophila melanogaster and its sibling species, D. simulans. Mol. Biol. Evol. 5, 182–191 (1988).
Makalowski, W. & Boguski, M. S. Evolutionary parameters of the transcribed mammalian genome: an analysis of 2,820 orthologous rodent and human sequences. Proc. Natl Acad. Sci. USA 95, 9407–9412 (1998).
Torgerson, D. G., Kulathinal, R. J. & Singh, R. S. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 19, 1973–1980 (2002).
Turner, L. M., Chuong, E. B. & Hoekstra, H. E. Comparative analysis of testis protein evolution in rodents. Genetics 179, 2075–2089 (2008).
Anderson, J. A., Gilliland, W. D. & Langley, C. H. Molecular population genetics and evolution of Drosophila meiosis genes. Genetics 181, 177–185 (2009).
Brand, C. L., Cattani, M. V., Kingan, S. B., Landeen, E. L. & Presgraves, D. C. Molecular evolution at a meiosis gene mediates species differences in the rate and patterning of recombination. Curr. Biol. 28, 1289–1295 (2018). This study uses phylogenetic and transgenic approaches to show that sequence differences in mei-218 between Drosophila species affect the function of the protein in controlling recombination rates.
Sidhu, G. K., Warzecha, T. & Pawlowski, W. P. Evolution of meiotic recombination genes in maize and teosinte. BMC Genomics 18, 106 (2017).
Dudka, D., Akins, R. B. & Lampson, M. A. FREEDA: An automated computational pipeline guides experimental testing of protein innovation. J. Cell Biol. 222, e202212084 (2023).
Bomblies, K., Higgins, J. D. & Yant, L. Meiosis evolves: adaptation to external and internal environments. N. Phytol. 208, 306–323 (2015).
Morgan, C. et al. Evolution of crossover interference enables stable autopolyploidy by ensuring pairwise partner connections in Arabidopsis arenosa. Curr. Biol. 31, 4713–4726 (2021). This study shows how increased strength of crossover interference can promote successful meiosis in new autotetraploid A. arenosa.
Morgan, C., Zhang, H., Henry, C. E., Franklin, F. C. H. & Bomblies, K. Derived alleles of two axis proteins affect meiotic traits in autotetraploid Arabidopsis arenosa. Proc. Natl Acad. Sci. USA 117, 8980–8988 (2020).
Melters, D. P., Paliulis, L. V., Korf, I. F. & Chan, S. W. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 20, 579–593 (2012).
Schvarzstein, M., Wignall, S. M. & Villeneuve, A. M. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 24, 219–228 (2010).
Barnes, T. M., Kohara, Y., Coulson, A. & Hekimi, S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141, 159–179 (1995).
Albertson, D. G., Rose, A. M. & Villeneuve, A. M. in C. elegans II. 2nd edn (eds Riddle, D. L. et al.) Ch. 3 (Cold Spring Harbor Laboratory Press, 1997).
Albertson, D. G. & Thomson, J. N. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res 1, 15–26 (1993).
Pan, J. et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731 (2011).
Kaback, D. B., Guacci, V., Barber, D. & Mahon, J. W. Chromosome size-dependent control of meiotic recombination. Science 256, 228–232 (1992).
Murakami, H. et al. Multilayered mechanisms ensure that short chromosomes recombine in meiosis. Nature 582, 124–128 (2020).
Acquaviva, L. et al. Ensuring meiotic DNA break formation in the mouse pseudoautosomal region. Nature 582, 426–431 (2020). This study describes how mouse sex chromosomes use a specialized mechanism to ensure recombination in the PAR and how a rapidly evolving protein, ANKRD31, is required.
Kauppi, L. et al. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331, 916–920 (2011).
Soriano, P. et al. High rate of recombination and double crossovers in the mouse pseudoautosomal region during male meiosis. Proc. Natl Acad. Sci. USA 84, 7218–7220 (1987).
Blackmon, H. & Demuth, J. P. The fragile Y hypothesis: Y chromosome aneuploidy as a selective pressure in sex chromosome and meiotic mechanism evolution. Bioessays 37, 942–950 (2015).
Ruiz-Herrera, A. & Waters, P. D. Fragile, unfaithful and persistent Ys — on how meiosis can shape sex chromosome evolution. Heredity 129, 22–30 (2022).
Papanikos, F. et al. Mouse ANKRD31 regulates spatiotemporal patterning of meiotic recombination initiation and ensures recombination between X and Y sex chromosomes. Mol. Cell 74, 1069–1085 (2019).
de la Fuente, R. et al. Meiotic pairing and segregation of achiasmate sex chromosomes in eutherian mammals: the role of SYCP3 protein. PLoS Genet. 3, e198 (2007).
Marin-Gual, L. et al. Strategies for meiotic sex chromosome dynamics and telomeric elongation in marsupials. PLoS Genet. 18, e1010040 (2022).
Haig, D. Games in tetrads: segregation, recombination, and meiotic drive. Am. Nat. 176, 404–413 (2010).
Werren, J. H. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl Acad. Sci. USA 108, 10863–10870 (2011).
Navarro-Dominguez, B. et al. Epistatic selection on a selfish segregation distorter supergene - drive, recombination, and genetic load. eLife 11, e78981 (2022).
Harvey, A. M. et al. A critical component of meiotic drive in Neurospora is located near a chromosome rearrangement. Genetics 197, 1165–1174 (2014).
Hu, W. et al. A large gene family in fission yeast encodes spore killers that subvert Mendel’s law. eLife 6, e26057 (2017).
Bravo Nunez, M. A., Sabbarini, I. M., Eide, L. E., Unckless, R. L. & Zanders, S. E. Atypical meiosis can be adaptive in outcrossed Schizosaccharomyces pombe due to wtf meiotic drivers. eLife 9, e57936 (2020). This study shows how increased levels of aneuploidy can be adaptive in the presence of a meiotic driver.
De Carvalho, M. et al. The wtf meiotic driver gene family has unexpectedly persisted for over 100 million years. eLife 11, e81149 (2022).
Lloyd, A., Morgan, C., FC, H. F. & Bomblies, K. Plasticity of meiotic recombination rates in response to temperature in Arabidopsis. Genetics 208, 1409–1420 (2018).
Kumar, S. et al. Timetree 5: an expanded resource for species divergence times. Mol. Biol. Evol. 39, msac174 (2022).
Lambing, C. et al. Differentiated function and localisation of SPO11-1 and PRD3 on the chromosome axis during meiotic DSB formation in Arabidopsis thaliana. PLoS Genet. 18, e1010298 (2022).
Zhang, Y. et al. Genetic interactions of histone modification machinery Set1 and PAF1C with the recombination complex Rec114-Mer2-Mei4 in the formation of meiotic DNA double-strand breaks. Int. J. Mol. Sci. 21, 2679 (2020).
Sun, W., Lorenz, A., Osman, F. & Whitby, M. C. A failure of meiotic chromosome segregation in a fbh1Delta mutant correlates with persistent Rad51–DNA associations. Nucleic Acids Res. 39, 1718–1731 (2011).
Joyce, E. F. et al. Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J. Cell Biol. 195, 359–367 (2011).
Li, Q., Hariri, S. & Engebrecht, J. Meiotic double-strand break processing and crossover patterning are regulated in a sex-specific manner by BRCA1-BARD1 in Caenorhabditis elegans. Genetics 216, 359–379 (2020).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Hong, S. et al. The logic and mechanism of homologous recombination partner choice. Mol. Cell 51, 440–453 (2013).
Kim, K. P. et al. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143, 924–937 (2010).
McMahill, M. S., Sham, C. W. & Bishop, D. K. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 5, e299 (2007).
Allers, T. & Lichten, M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol. Cell 8, 225–231 (2001).
Hunter, N. & Kleckner, N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70 (2001).
Schwacha, A. & Kleckner, N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83, 783–791 (1995).
Paigen, K. & Petkov, P. M. PRDM9 and its role in genetic recombination. Trends Genet. 34, 291–300 (2018).
Grey, C., Baudat, F. & de Massy, B. PRDM9, a driver of the genetic map. PLoS Genet. 14, e1007479 (2018).
Gregorova, S. et al. Modulation of Prdm9-controlled meiotic chromosome asynapsis overrides hybrid sterility in mice. eLife 7, e34282 (2018).
Baker, Z., Przeworski, M. & Sella, G. Down the Penrose stairs, or how selection for fewer recombination hotspots maintains their existence. eLife 12, e83769 (2023).
Davies, B. et al. Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530, 171–176 (2016). This study shows that a humanized PRDM9 allele can rescue hybrid sterility in mice by re-directing meiotic DSB hot spots.
Zelkowski, M., Olson, M. A., Wang, M. & Pawlowski, W. Diversity and determinants of meiotic recombination landscapes. Trends Genet. 35, 359–370 (2019).
Henderson, I. R. & Bomblies, K. Evolution and plasticity of genome-wide meiotic recombination rates. Annu. Rev. Genet. 55, 23–43 (2021). This review covers the literature that describes the variation of meiotic recombination rates.
Acknowledgements
The authors apologize to authors whose work could not be cited owing to space limitations. They thank M. Marcet-Ortega for the micrograph of the mouse spermatocyte spread in Fig. 1b. M.A. was supported by an EMBO long-term fellowship (ALTF 905-2019) and a postdoctoral award from the SKI Basic Research Innovation Award Initiative and the Dewitt Wallace Basic Science Award Fund. Research in the Keeney lab is supported in part by National Cancer Institute Cancer Center support grant P30 CA08748, NIH grants R35 GM118092 (to S.K.) and R01 HD110120 (to S.K. and D. Patel) and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information:
Nature Reviews Genetics thanks Ian Henderson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Aneuploidy
-
Describes the occurrence of missing or extra chromosomes (or parts of chromosomes) in a cell. Meiotic errors that lead to aneuploidy in gametes can have detrimental consequences for an organism.
- Crossover
-
One potential product of the homologous recombination DNA repair pathway. During a crossover event, the DNA flanking the break site is reciprocally exchanged.
- Crossover interference
-
Describes the observation that crossovers show a non-random distribution along chromosomes in which presence of a crossover is accompanied by a reduced likelihood of finding another crossover nearby. As a consequence of interference, crossovers tend to be widely and evenly spaced.
- DNA double-strand breaks
-
DNA lesions in which both DNA strands are cut. During meiosis, DNA double-strand breaks are essential to initiate homologous recombination.
- Haploidization
-
The reduction of the genome complement by half. Haploidization is essential during meiosis to allow the restoration of the genome content at fertilization.
- Homologous chromosomes
-
Homologous chromosomes (homologues) in diploid organisms are pairs of chromosomes that contain the same genes but are of different parental origin. During meiosis, homologous chromosomes are paired to achieve segregation.
- Meiotic drivers
-
Genetic loci that gain a transmission advantage, not by conferring a fitness advantage, but instead by promoting their inheritance in more than 50% of gametes through manipulation of the meiotic process.
- Molecular evolution
-
The study of evolutionary changes at the DNA level. The molecular evolution approaches discussed in this Review aim to distinguish whether protein-coding genes or gene domains experience purifying or positive selection.
- Non-crossover
-
A product of homologous recombination in which no reciprocal exchange of chromosome arms happens, and exchange of genetic information (usually non-reciprocal) is limited.
- Positive selection
-
Describes a process in which novel genetic variants confer a fitness advantage and thus become fixed in a population. This is in contrast to genetic variants that are neutral and become fixed owing to genetic drift.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Arter, M., Keeney, S. Divergence and conservation of the meiotic recombination machinery. Nat Rev Genet 25, 309–325 (2024). https://doi.org/10.1038/s41576-023-00669-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-023-00669-8