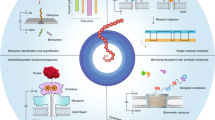
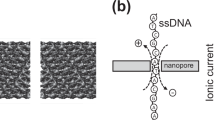
Abstract
Sequencing of nucleic acids with nanopores has emerged as a powerful tool offering rapid readout, high accuracy, low cost and portability. This label-free method for sequencing at the single-molecule level is an achievement on its own. However, nanopores also show promise for the technologically even more challenging sequencing of polypeptides, something that could considerably benefit biological discovery, clinical diagnostics and homeland security, as current techniques lack portability and speed. Here we survey the biochemical innovations underpinning commercial and academic nanopore DNA/RNA sequencing techniques, and explore how these advances can fuel developments in future protein sequencing with nanopores.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Logsdon, G. A., Vollger, M. R. & Eichler, E. E. Long-read human genome sequencing and its applications. Nat. Rev. Genet. 21, 597–614 (2020).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Charalampous, T. et al. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Nat. Biotechnol. 37, 783–792 (2019).
Deng, X. et al. Metagenomic sequencing with spiked primer enrichment for viral diagnostics and genomic surveillance. Nat. Microbiol. 5, 443–454 (2020).
Zhou, B. et al. SARS-CoV-2 spike D614G change enhances replication and transmission. Nature 592, 122–127 (2021).
Cao, X. et al. Alt-RPL36 downregulates the PI3K-AKT-mTOR signaling pathway by interacting with TMEM24. Nat. Commun. 12, 508 (2021).
Liu, Y. et al. Proteomic profiling of HIV-1 infection of human CD4+ T cells identifies PSGL-1 as an HIV restriction factor. Nat. Microbiol. 4, 813–825 (2019).
Leggett, R. M. et al. Rapid MinION profiling of preterm microbiota and antimicrobial-resistant pathogens. Nat. Microbiol. 5, 430–442 (2020).
Li, D. et al. Genomic profiling informs diagnoses and treatment in vascular anomalies. Nat. Med. 29, 1530–1539 (2023).
Macken, W. L., Vandrovcova, J., Hanna, M. G. & Pitceathly, R. D. S. Applying genomic and transcriptomic advances to mitochondrial medicine. Nat. Rev. Neurol. 17, 215–230 (2021).
Sero, D. et al. Facial recognition from DNA using face-to-DNA classifiers. Nat. Commun. 10, 2557 (2019).
Cornelis, S., Gansemans, Y., Deleye, L., Deforce, D. & Van Nieuwerburgh, F. Forensic SNP genotyping using nanopore MinION sequencing. Sci. Rep. 7, 41759 (2017).
Liao, J. et al. Nationwide genomic atlas of soil-dwelling Listeria reveals effects of selection and population ecology on pangenome evolution. Nat. Microbiol. 6, 1021–1030 (2021).
Cuypers, W. L. et al. A global genomic analysis of Salmonella Concord reveals lineages with high antimicrobial resistance in Ethiopia. Nat. Commun. 14, 3517 (2023).
Eid, J. et al. Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009).
Bentley, D. R. et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456, 53–59 (2008).
Booth, M. J. in Nucleic Acids in Chemistry and Biology (eds Blackburn, G. M. et al.) Ch. 8 (The Royal Society of Chemistry, 2022).
Eisenstein, M. Illumina faces short-read rivals. Nat. Biotechnol. 41, 3–5 (2023).
Arslan, S. et al. Sequencing by avidity enables high accuracy with low reagent consumption. Nat. Biotechnol. 42, 132–138 (2024).
Rothberg, J. M. et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 475, 348–352 (2011).
Slatko, B. E., Gardner, A. F. & Ausubel, F. M. Overview of next-generation sequencing technologies. Curr. Protoc. Mol. Biol. 122, e59 (2018).
Quick, J. et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 530, 228–232 (2016).
Jain, M. et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018).
Wang, Y. Y., Zhao, Y., Bollas, A., Wang, Y. Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Marx, V. Method of the year: long-read sequencing. Nat. Methods 20, 6–11 (2023).
Deamer, D., Akeson, M., Branton, D., Kasianowicz, J. J. & Bezrukov, S. M. Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524 (2016).
Wanunu, M. Nanopores: a journey towards DNA sequencing. Phys. Life Rev. 9, 125–158 (2012).
Alper, J. From the bioweapons trenches, new tools for battling microbes. Science 284, 1754–1755 (1999).
Kasianowicz, J. J. & Bezrukov, S. M. On ‘three decades of nanopore sequencing’. Nat. Biotechnol. 34, 481–482 (2016).
Bayley, H. Nanopore sequencing: from imagination to reality. Clin. Chem. 61, 25–31 (2015).
Liu, Y. et al. DNA methylation-calling tools for Oxford Nanopore sequencing: a survey and human epigenome-wide evaluation. Genome Biol. 22, 295 (2021).
Yuen, Z. W. et al. Systematic benchmarking of tools for CpG methylation detection from nanopore sequencing. Nat. Commun. 12, 3438 (2021).
Lu, H., Giordano, F. & Ning, Z. Oxford nanopore MinION sequencing and genome assembly. Genom. Proteom. Bioinform. 14, 265–279 (2016).
Brinkerhoff, H., Kang, A. S. W., Liu, J., Aksimentiev, A. & Dekker, C. Multiple rereads of single proteins at single-amino acid resolution using nanopores. Science 374, 1509–1513 (2021).
Yan, S. et al. Single molecule ratcheting motion of peptides in a Mycobacterium smegmatis Porin A (MspA) nanopore. Nano Lett. 21, 6703–6710 (2021).
Nivala, J., Marks, D. B. & Akeson, M. Unfoldase-mediated protein translocation through an α-hemolysin nanopore. Nat. Biotechnol. 31, 247–250 (2013).
Zhang, S. et al. Bottom-up fabrication of a proteasome-nanopore that unravels and processes single proteins. Nat. Chem. 13, 1192–1199 (2021).
Ouldali, H. et al. Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore. Nat. Biotechnol. 38, 176–181 (2020).
Motone, K. et al. Multi-pass, single-molecule nanopore reading of long protein strands with single-amino acid sensitivity. Preprint at https://doi.org/10.1101/2023.10.19.563182 (2023).
Martin-Baniandres, P. et al. Enzyme-less nanopore detection of post-translational modifications within long polypeptides. Nat. Nanotechnol 18, 1335–1340 (2023).
Sauciuc, A. et al. Translocation of linearized full-length proteins through an engineered nanopore under opposing electrophoretic force. Nat Biotechnol. https://doi.org/10.1038/s41587-023-01954-x (2023).
Zhang, Y. et al. Peptide sequencing based on host–guest interaction-assisted nanopore sensing. Nat. Methods 21, 102–109 (2024).
Wang, K. et al. Unambiguous discrimination of all 20 proteinogenic amino acids and their modifications by nanopore. Nat. Methods 21, 92–101 (2024).
Drachman, N. Nanopore ion sources deliver single amino acid and peptide ions directly into high vacuum. Preprint at https://www.researchsquare.com/article/rs-1686064/v1 (2022).
Alfaro, J. A. et al. The emerging landscape of single-molecule protein sequencing technologies. Nat. Methods 18, 604–617 (2021).
Eisenstein, M. Seven technologies to watch in 2023. Nature 613, 794–797 (2023).
MacCoss, M. J., Alfaro, J., Wanunu, M., Faivre, D. A. & Slavov, N. Sampling the proteome by emerging single-molecule and mass-spectrometry methods. Nat. Methods 20, 339–346 (2022).
Leggett, R. M. & Clark, M. D. A world of opportunities with nanopore sequencing. J. Exp. Bot. 68, 5419–5429 (2017).
Brown, C. G. & Clarke, J. Nanopore development at Oxford Nanopore. Nat. Biotechnol. 34, 810–811 (2016).
Restrepo-Pérez, L., Joo, C. & Dekker, C. Paving the way to single-molecule protein sequencing. Nat. Nanotechnol. 13, 786–796 (2018).
Howorka, S. Building membrane nanopores. Nat. Nanotechnol. 12, 619–630 (2017).
Feng, J. et al. Identification of single nucleotides in MoS2 nanopores. Nat. Nanotechnol. 10, 1070–1076 (2015).
Miles, B. N. et al. Single molecule sensing with solid-state nanopores: novel materials, methods and applications. Chem. Soc. Rev. 42, 15–28 (2013).
Yusko, E. C. et al. Real-time shape approximation and fingerprinting of single proteins using a nanopore. Nat. Nanotechnol. 12, 360–367 (2017).
Wei, R., Gatterdam, V., Wieneke, R., Tampé, R. & Rant, U. Stochastic sensing of proteins with receptor-modified solid-state nanopores. Nat. Nanotechnol. 7, 257–263 (2012).
Dekker, C. Solid-state nanopores. Nat. Nanotechnol. 2, 209–215 (2007).
Tunuguntla, R. H. et al. Enhanced water permeability and tunable ion selectivity in subnanometer carbon nanotube porins. Science 359, 792–796 (2018).
He, Y., Tsutsui, M., Zhou, Y. & Miao, X. S. Solid-state nanopore systems: from materials to applications. NPG Asia Mater. 13, 48 (2021).
Xue, L. et al. Solid-state nanopore sensors. Nat. Rev. Mater. 5, 931–951 (2020).
Moreau, C. J., Dupuis, J. P., Revilloud, J., Arumugam, K. & Vivaudou, M. Coupling ion channels to receptors for biomolecule sensing. Nat. Nanotechnol. 3, 620–625 (2008).
Cao, C. et al. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 11, 713–718 (2016).
Soskine, M. et al. An engineered ClyA nanopore detects folded target proteins by selective external association and pore entry. Nano Lett. 12, 4895–4900 (2012).
Song, L. et al. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science 274, 1859–1866 (1996).
Faller, M., Niederweis, M. & Schulz, G. E. The structure of a mycobacterial outer-membrane channel. Science 303, 1189–1192 (2004).
Clarke, J. et al. Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol. 4, 265–270 (2009).
Manrao, E. A. et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 30, 349–353 (2012).
Cherf, G. M. et al. Automated forward and reverse ratcheting of DNA in a nanopore at five angstrom precision. Nat. Biotechnol. 30, 344–348 (2012).
Stoddart, D. et al. Nucleobase recognition in ssDNA at the central constriction of the α-hemolysin pore. Nano Lett. 10, 3633–3637 (2010).
Stoddart, D., Maglia, G., Mikhailova, E., Heron, A. J. & Bayley, H. Multiple base-recognition sites in a biological nanopore: two heads are better than one. Angew. Chem. Int. Ed. 49, 556–559 (2010).
Van Gerven, N., Van der Verren, S. E., Reiter, D. M. & Remaut, H. The role of functional amyloids in bacterial virulence. J. Mol. Biol. 430, 3657–3684 (2018).
Goyal, P. et al. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516, 250–253 (2014).
Cao, B. et al. Structure of the nonameric bacterial amyloid secretion channel. Proc. Natl Acad. Sci. USA 111, E5439–E5444 (2014).
Boža, V., Brejová, B. & Vinař, T. DeepNano: deep recurrent neural networks for base calling in MinION nanopore reads. PLoS ONE 12, e0178751 (2017).
David, M., Dursi, L. J., Yao, D., Boutros, P. C. & Simpson, J. T. Nanocall: an open source basecaller for Oxford nanopore sequencing data. Bioinformatics. 33, 49–55 (2017).
Jayasinghe, L. & Wallace, J. E. Mutant pore. US patent 11186868 B2 (2021).
Nanopore Sequencing Accuracy. Oxford Nanopore Technologies https://nanoporetech.com/accuracy (2023).
Chapman, M. R. et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855 (2002).
Sereika, M. et al. Oxford Nanopore R10.4 long-read sequencing enables the generation of near-finished bacterial genomes from pure cultures and metagenomes without short-read or reference polishing. Nat. Methods 19, 823–826 (2022).
Kasianowicz, J. J., Brandin, E., Branton, D. & Deamer, D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl Acad. Sci. USA 93, 13770–13773 (1996).
Meller, A., Nivon, L. & Branton, D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 86, 3435–3438 (2001).
Laszlo, A. H., Derrington, I. M. & Gundlach, J. H. MspA nanopore as a single-molecule tool: from sequencing to SPRNT. Methods 105, 75–89 (2016).
Heron, A. et al. Modified helicases. US patent 2018/0037874 A9 (2018).
He, X. et al. The T4 phage SF1B helicase Dda Is structurally optimized to perform DNA strand separation. Structure 20, 1189–1200 (2012).
Craig, J. M. et al. Revealing dynamics of helicase translocation on single-stranded DNA using high-resolution nanopore tweezers. Proc. Natl Acad. Sci. USA 114, 11932–11937 (2017).
Stava, E. et al. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat. Biotechnol. 33, 1073–1075 (2015).
Noakes, M. T. et al. Increasing the accuracy of nanopore DNA sequencing using a time-varying cross membrane voltage. Nat. Biotechnol. 37, 651–656 (2019).
Laszlo, A. H., Derrrington, I. M. & Gundlach, J. H. Subangstrom measurements of enzyme function using a biological nanopore, SPRNT. Methods Enzymol. 582, 387–414 (2017).
Jain, M., Olsen, H. E., Paten, B. & Akeson, M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 17, 239 (2016).
Clarke, J., White, J., Milton, J. & Brown, C. Coupling method. PCT patent 2012/164270 A1 (2012).
Stark, R., Grzelak, M. & Hadfield, J. RNA sequencing: the teenage years. Nat. Rev. Genet. 20, 631–656 (2019).
Byrne, A. et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 8, 16027 (2017).
Garalde, D. R. et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 15, 201–206 (2018).
Workman, R. E. et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 16, 1297–1305 (2019).
Abu-Shumays, R. L. et al. Direct nanopore sequencing of individual full length tRNA strands. ACS Nano 15, 16642–16653 (2021).
Olsen, H.E. et al. Advances in nanopore direct RNA sequencing. Nat. Methods 19, 1160–1164 (2022).
Miller, C. Ion Channel Reconstitution (Springer, 2013).
Cao, C., Liao, D. F., Yu, J., Tian, H. & Long, Y. T. Construction of an aerolysin nanopore in a lipid bilayer for single-oligonucleotide analysis. Nat. Protoc. 12, 1901–1911 (2017).
Baba, T. et al. Formation and characterization of planar lipid bilayer membranes from synthetic phytanyl-chained glycolipids. Biochim. Biophys. Acta Biomembr. 1421, 91–102 (1999).
Schmidt, J. Membrane platforms for biological nanopore sensing and sequencing. Curr. Opin. Biotechnol. 39, 17–27 (2016).
Holden, M. A., Needham, D. & Bayley, H. Functional bionetworks from nanoliter water droplets. J. Am. Chem. Soc. 129, 8650–8655 (2007).
Funakoshi, K., Suzuki, H. & Takeuchi, S. Lipid bilayer formation by contacting monolayers in a microfluidic device for membrane protein analysis. Anal. Chem. 78, 8169–8174 (2006).
Urban, M. et al. Highly parallel transport recordings on a membrane-on-nanopore chip at single molecule resolution. Nano Lett. 14, 1674–1680 (2014).
Jeon, T.-J., Malmstadt, N. & Schmidt, J. J. Hydrogel-encapsulated lipid membranes. J. Am. Chem. Soc. 128, 42–43 (2006).
Shim, J. W. & Gu, L. Q. Stochastic sensing on a modular chip containing a single-ion channel. Anal. Chem. 79, 2207–2213 (2007).
Heitz, B. A., Jones, I. W., Hall, H. K., Aspinwall, C. A. & Saavedra, S. S. Fractional polymerization of a suspended planar bilayer creates a fluid, highly stable membrane for ion channel recordings. J. Am. Chem. Soc. 132, 7086–7093 (2010).
Daly, S. M., Heffernan, L. A., Barger, W. R. & Shenoy, D. K. Photopolymerization of mixed monolayers and black lipid membranes containing gramicidin A and diacetylenic phospholipids. Langmuir 22, 1215–1222 (2006).
Nardin, C., Winterhalter, M. & Meier, W. Giant free-standing ABA triblock copolymer membranes. Langmuir 16, 7708–7712 (2000).
Meier, W., Nardin, C. & Winterhalter, M. Reconstitution of channel proteins in (polymerized) ABA triblock copolymer membranes. Angew. Chem. Int. Ed. 39, 4599–4602 (2000).
Hyde, J. R. et al. Formation of array of membranes and apparatus therefor. WO2014064443A3 (2014).
González-Pérez, A., Stibius, K. B., Vissing, T., Nielsen, C. H. & Mouritsen, O. G. Biomimetic triblock copolymer membrane arrays: a stable template for functional membrane proteins. Langmuir 25, 10447–10450 (2009).
De Coster, W., D’Hert, S., Schultz, D. T., Cruts, M. & Van Broeckhoven, C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 34, 2666–2669 (2018).
Fukasawa, Y., Ermini, L., Wang, H., Carty, K. & Cheung, M.-S. LongQC: a quality control tool for third generation sequencing long read data. G3 Genes Genomes Genet. 10, 1193–1196 (2020).
Lanfear, R., Schalamun, M., Kainer, D., Wang, W. & Schwessinger, B. MinIONQC: fast and simple quality control for MinION sequencing data. Bioinformatics. 35, 523–525 (2019).
Tarraga, J., Gallego, A., Arnau, V., Medina, I. & Dopazo, J. HPG pore: an efficient and scalable framework for nanopore sequencing data. BMC Bioinformatics 17, 107 (2016).
Loman, N. J. & Quinlan, A. R. Poretools: a toolkit for analyzing nanopore sequence data. Bioinformatics 30, 3399–3401 (2014).
Payne, A., Holmes, N., Rakyan, V. & Loose, M. BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 35, 2193–2198 (2019).
Gamaarachchi, H. et al. Fast nanopore sequencing data analysis with SLOW5. Nat. Biotechnol. 40, 1026–1029 (2022).
Simpson, J. T. et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods 14, 407–410 (2017).
Liu, Q. et al. Detection of DNA base modifications by deep recurrent neural network on Oxford Nanopore sequencing data. Nat. Commun. 10, 2449 (2019).
Ni, P. et al. DeepSignal: detecting DNA methylation state from Nanopore sequencing reads using deep-learning. Bioinformatics 35, 4586–4595 (2019).
Liu, Q. et al. NanoMod: a computational tool to detect DNA modifications using nanopore long-read sequencing data. BMC Genomics 20, 78 (2019).
Liu, H. et al. Accurate detection of m6A RNA modifications in native RNA sequences. Nat. Commun. 10, 4079 (2019).
Pratanwanich, P. N. et al. Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore. Nat. Biotechnol. 39, 1394–1402 (2021).
Leger, A. et al. RNA modifications detection by comparative Nanopore direct RNA sequencing. Nat. Commun. 12, 7198 (2021).
Hendra, C. et al. Detection of m6A from direct RNA sequencing using a multiple instance learning framework. Nat. Methods 19, 1590–1598 (2022).
Miclotte, G. et al. Jabba: hybrid error correction for long sequencing reads. Algorithms Mol. Biol. 11, 10 (2016).
Morisse, P., Lecroq, T. & Lefebvre, A. Hybrid correction of highly noisy long reads using a variable-order de Bruijn graph. Bioinformatics 34, 4213–4222 (2018).
Madoui, M.-A. et al. Genome assembly using Nanopore-guided long and error-free DNA reads. BMC Genomics 16, 327 (2015).
Sović, I. et al. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat. Commun. 7, 11307 (2016).
Sedlazeck, F. J. et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods 15, 461–468 (2018).
Li, H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110 (2016).
Ewing, A. D. et al. Nanopore sequencing enables comprehensive transposable element epigenomic profiling. Mol. Cell 80, 915–928.e5 (2020).
Deonovic, B., Wang, Y., Weirather, J., Wang, X.-J. & Au, K. F. IDP-ASE: haplotyping and quantifying allele-specific expression at the gene and gene isoform level by hybrid sequencing. Nucleic Acids Res. 45, e32 (2017).
Lucká, M. et al. WarpSTR: determining tandem repeat lengths using raw nanopore signals. Bioinformatics 39, btad388 (2023).
Mohamed, M. et al. TrEMOLO: accurate transposable element allele frequency estimation using long-read sequencing data combining assembly and mapping-based approaches. Genome Biol. 24, 63 (2023).
De Lannoy, C. et al. The long reads ahead: de novo genome assembly using the MinION. F1000Res. 6, 1083 (2017).
Anderson, B. N. Optically based nanopore sequencing. EP patent 3482196 B1 (2022).
Huber, M. & Davidson, S. Efficient optical analysis of polymers using arrays of nanostructures. EP patent 3209800 B1 (2022).
Huber, M. Nanopore-based polymer analysis with mutually-quenching fluorescent labels. WO2016057829A1 (2016).
Huber, M., Assad, O., Cleek, T. & Davlieva, M. Fluorescent polynucleotide sequencing methods and compositions. WO20230193376 (2023).
Buzby, P. R., Meller, A., McNally, B., Fan, A. & Olejnik-Krzynmanska, E. Sequence preserved DNA conversion for optical nanopore sequencing. WO2012135658A2 (2012).
McNally, B. et al. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 10, 2237–2244 (2010).
Kokoris, M. S. & McRuer, R. N. High throughput nucleic acid sequencing by expansion. US patent US7939259B2 (2011).
Kumar, S. et al. PEG-labeled nucleotides and nanopore detection for single molecule DNA sequencing by synthesis. Sci. Rep. 2, 684 (2012).
Masoud, V. et al. Systems and methods for measurment and sequencing of bio-molecules. EP patent 3449247 B1 (2021).
Ohshiro, T. et al. Single-molecule electrical random resequencing of DNA and RNA. Sci. Rep. 2, 501 (2012).
Tsutsui, M., Taniguchi, M., Yokota, K. & Kawai, T. Identifying single nucleotides by tunnelling current. Nat. Nanotechnol. 5, 286–290 (2010).
Lagerqvist, J., Zwolak, M. & Di Ventra, M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 6, 779–782 (2006).
Huang, S. et al. Identifying single bases in a DNA oligomer with electron tunnelling. Nat. Nanotechnol. 5, 868–873 (2010).
Lee, J. W. Nanoelectrode-gated detection of individual molecules with potential for rapid DNA sequencing. Solid State Phenom. 121–123, 1379–1386 (2007).
Zhang, Y. et al. Single molecule DNA resensing using a two-pore device. Small 14, 1801890 (2018).
Liu, X., Zhang, Y., Nagel, R., Reisner, W. & Dunbar, W. B. Controlling DNA tug-of-war in a dual nanopore device. Small 15, 1901704 (2019).
Liu, X. et al. Flossing DNA in a dual nanopore device. Small 16, 1905379 (2020).
Rand, A. et al. Electronic mapping of a bacterial genome with dual solid-state nanopores and active single-molecule control. ACS Nano 16, 5258–5273 (2021).
Choudhary, A. et al. High-fidelity capture, threading, and infinite-depth sequencing of single DNA molecules with a double-nanopore system. ACS Nano. 14, 15566–15576 (2020).
Oliver, J. S. Devices and methods for determining the length of biopolymers and distances between probes bound thereto. US patent 8262879 B2 (2012).
Oliver, J. S. et al. High-definition electronic genome maps from single molecule data. Preprint at https://www.biorxiv.org/content/10.1101/139840v1 (2017).
Passera, A. et al. Characterization of Lysinibacillus fusiformis strain S4C11: in vitro, in planta and in silico analyses reveal a plant-benefcial microbe. Microbiol. Res. 244, 126665 (2021).
Weigand, M. R. et al. Screening and genomic characterization of filamentous hemagglutinin-deficient Bordetella pertussis. Infect. Immun. 86, 5–7 (2018).
Kaiser, M. D. et al. Automated structural variant verification in human genomes using single-molecule electronic DNA mapping. Preprint at https://www.biorxiv.org/content/10.1101/140699v1 (2017).
Bošković, F. et al. Simultaneous identification of viruses and viral variants with programmable DNA nanobait. Nat. Nanotechnol. 18, 290–298 (2023).
Bošković, F. & Keyser, U. F. Nanopore microscope identifies RNA isoforms with structural colours. Nat. Chem. 14, 1258–1264 (2022).
Takulapalli, B. Field effect transistor, device including the transistor, and methods of forming and using same. US patent 2020/0096505 A1 (2020).
Li, R. et al. QitanTech nanopore long-read sequencing enables rapid resolution of complete genomes of multi-drug resistant pathogens. Front. Microbiol. 13, 778659 (2022).
Hou, Y. et al. Forensic nanopore sequencing of microhaplotype markers using QitanTech’s QNome. Genetics 57, 102657 (2022).
Yam, C. et al. DNA sequencing based on electronic tunneling in a gold nanogap: a first-principles study. Phys. Chem. Chem. Phys. 24, 5748–5754 (2022).
Traversi, F. et al. Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nat. Nanotechnol. 8, 939–945 (2013).
Mojtabavi, M., Vahidmohammadi, A., Liang, W., Beidaghi, M. & Wanunu, M. Single-molecule sensing using nanopores in two-dimensional transition metal carbide (MXene) membranes. ACS Nano. 13, 3042–3053 (2019).
Schneider, G. F. et al. Tailoring the hydrophobicity of graphene for its use as nanopores for DNA translocation. Nat. Commun. 4, 2619 (2013).
Nicoli, F., Verschueren, D., Klein, M., Dekker, C. & Jonsson, M. P. DNA translocations through solid-state plasmonic nanopores. Nano Lett. 14, 6917–6925 (2014).
Assad, O. N. et al. Light-enhancing plasmonic-nanopore biosensor for superior single-molecule detection. Adv. Mater. 29, 1605442 (2017).
Larkin, J., Henley, R. Y., Jadhav, V., Korlach, J. & Wanunu, M. Length-independent DNA packing into nanopore zero-mode waveguides for low-input DNA sequencing. Nat. Nanotechnol. 12, 1169–1175 (2017).
Zhang, Y., Tang, Y., Tan, C. & Xu, W. Toward nanopore electrospray mass spectrometry: nanopore effects in the analysis of bacteria. ACS Cent. Sci. 6, 1001–1008 (2020).
Bush, J. et al. The nanopore mass spectrometer. Rev. Sci. Instrum. 88, 113307 (2017).
Movileanu, L., Howorka, S., Braha, O. & Bayley, H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 18, 1091–1095 (2001).
Thakur, A. K. & Movileanu, L. Real-time measurement of protein–protein interactions at single-molecule resolution using a biological nanopore. Nat. Biotechnol. 37, 96–104 (2019).
Mayer, S. F., Cao, C. & Peraro, M. D. Biological nanopores for single-molecule sensing. iScience 25, 104145 (2022).
Lukoyanova, N. et al. Conformational changes during pore formation by the perforin-related protein pleurotolysin. PLoS Biol. 13, 1002049 (2015).
Wallace, A. J. et al. E. coli hemolysin E (Hlye, ClyA, SheA): X-ray crystal structure of the toxin and observation of membrane pores by electron microscopy. Cell 100, 265–276 (2000).
Huang, G. et al. Electro-osmotic vortices promote the capture of folded proteins by PlyAB nanopores. Nano Lett. 20, 3819–3827 (2020).
Galenkamp, N. S., Soskine, M., Hermans, J., Wloka, C. & Maglia, G. Direct electrical quantification of glucose and asparagine from bodily fluids using nanopores. Nat. Commun. 9, 4085 (2018).
Soskine, M., Biesemans, A., De Maeyer, M. & Maglia, G. Tuning the size and properties of ClyA nanopores assisted by directed evolution. J. Am. Chem. Soc. 135, 13456–13463 (2013).
Galenkamp, N. S., Biesemans, A. & Maglia, G. Directional conformer exchange in dihydrofolate reductase revealed by single-molecule nanopore recordings. Nat. Chem. 12, 481–488 (2020).
Xing, Y., Dorey, A., Jayasinghe, L. & Howorka, S. Highly shape- and size-tunable membrane nanopores made with DNA. Nat. Nanotechnol. 17, 708–713 (2022).
Lanphere, C. et al. Design, assembly and characterization of membrane-spanning DNA nanopores. Nat. Protoc. 16, 86–130 (2021).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Seeman, N. C. & Sleiman, H. F. DNA nanotechnology. Nat. Rev. Mater. 3, 17068 (2017).
Jiang, S., Ge, Z., Mou, S., Yan, H. & Fan, C. Designer DNA nanostructures for therapeutics. Chem 7, 1156–1179 (2021).
Dey, S. et al. A reversibly gated protein-transporting membrane channel made of DNA. Nat. Commun. 13, 2271 (2022).
Diederichs, T. et al. Synthetic protein-conductive membrane nanopores built with DNA. Nat. Commun. 10, 5018 (2019).
Kwok, H., Briggs, K. & Tabard-Cossa, V. Nanopore fabrication by controlled dielectric breakdown. PLoS ONE 9, e92880 (2014).
Li, W. et al. Single protein molecule detection by glass nanopores. ACS Nano 7, 4129–4134 (2013).
Bell, N. A. W., Chen, K., Ghosal, S., Ricci, M. & Keyser, U. F. Asymmetric dynamics of DNA entering and exiting a strongly confining nanopore. Nat. Commun. 8, 380 (2017).
Sze, J. Y. Y., Ivanov, A. P., Cass, A. E. G. & Edel, J. B. Single molecule multiplexed nanopore protein screening in human serum using aptamer modified DNA carriers. Nat. Commun. 8, 1552 (2017).
Maffeo, C. et al. Modeling and simulation of ion channels. Chem. Rev. 112, 6250–6284 (2012).
Varongchayakul, N., Song, J., Meller, A. & Grinstaff, M. W. Single-molecule protein sensing in a nanopore: a tutorial. Chem. Soc. Rev. 47, 8512–8524 (2018).
Firnkes, M., Pedone, D., Knezevic, J., Döblinger, M. & Rant, U. Electrically facilitated translocations of proteins through silicon nitride nanopores: conjoint and competitive action of diffusion, electrophoresis and electroosmosis. Nano Lett. 10, 2162–2167 (2010).
Schmid, S., Stömmer, P., Dietz, H. & Dekker, C. Nanopore electro-osmotic trap for the label-free study of single proteins and their conformations. Nat. Nanotechnol. 16, 1244–1250 (2021).
Bandara, Y. M. N. D. Y., Farajpour, N. & Freedman, K. J. Nanopore current enhancements lack protein charge dependence and elucidate maximum unfolding at protein’s isoelectric point. J. Am. Chem. Soc. 144, 3063–3073 (2022).
Restrepo-Pérez, L., Shalini, J., Aksimentiev, A., Joo, C. & Dekker, C. SDS-assisted protein transport through solid-state nanopores. Nanoscale 9, 11685–11693 (2017).
Rivas, F. et al. Label-free analysis of physiological hyaluronan size distribution with a solid-state nanopore sensor. Nat. Commun. 9, 1037 (2018).
Plesa, C. et al. Fast translocation of proteins through solid state nanopores. Nano Lett. 13, 658–663 (2013).
Lin, C. Y. et al. Ultrafast polymer dynamics through a nanopore. Nano Lett. 22, 8719–8727 (2022).
Chau, C. C., Radford, S. E., Hewitt, E. W. & Actis, P. Macromolecular crowding enhances the detection of DNA and proteins by a solid-state nanopore. Nano Lett. 20, 5553–5561 (2020).
Keyser, U. F. Enhancing nanopore sensing with DNA nanotechnology. Nat. Nanotechnol. 11, 106–108 (2016).
Bell, N. A. W. & Keyser, U. F. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotechnol. 11, 645–651 (2016).
Tripathi, P. et al. Electrical unfolding of cytochrome c during translocation through a nanopore constriction. Proc. Natl Acad. Sci. USA 118, e2016262118 (2021).
Movileanu, L., Schmittschmitt, J. P., Scholtz, J. M. & Bayley, H. Interactions of peptides with a protein pore. Biophys. J. 89, 1030–1045 (2005).
Rodriguez-Larrea, D. & Bayley, H. Multistep protein unfolding during nanopore translocation. Nat. Nanotechnol. 8, 288–295 (2013).
Feng, J. et al. Transmembrane protein rotaxanes reveal kinetic traps in the refolding of translocated substrates. Commun. Biol. 3, 159 (2020).
Rosen, C. B., Bayley, H. & Rodriguez-Larrea, D. Free-energy landscapes of membrane co-translocational protein unfolding. Commun. Biol. 3, 160 (2020).
Rodriguez-Larrea, D. Single-amino acid discrimination in proteins with homogeneous nanopore sensors and neural networks. Biosens. Bioelectron. 180, 113108 (2021).
Nivala, J., Mulroney, L., Li, G., Schreiber, J. & Akeson, M. Discrimination among protein variants using an unfoldase-coupled nanopore. ACS Nano 8, 12365–12375 (2014).
Yao, Y., Docter, M., Van Ginkel, J., De Ridder, D. & Joo, C. Single-molecule protein sequencing through fingerprinting: computational assessment. Phys. Biol. 12, 055003 (2015).
Ohayon, S., Girsault, A., Nasser, M., Shen-Orr, S. & Meller, A. Simulation of single-protein nanopore sensing shows feasibility for whole-proteome identification. PLoS Comput. Biol. 15, e1007067 (2019).
Lucas, F. L. R., Versloot, R. C. A., Yakovlieva, L., Walvoort, M. T. C. & Maglia, G. Protein identification by nanopore peptide profiling. Nat. Commun. 12, 5795 (2021).
Afshar Bakshloo, M. et al. Nanopore-based protein identification. J. Am. Chem. Soc. 144, 2716–2725 (2022).
Yu, L. et al. Unidirectional single-file transport of full-length proteins through a nanopore. Nat. Biotechnol. 41, 1130–1139 (2023).
Restrepo-Pérez, L. et al. Resolving chemical modifications to a single amino acid within a peptide using a biological nanopore. ACS Nano 13, 13668–13676 (2019).
Wang, R. et al. Single-molecule discrimination of labeled DNAs and polypeptides using photoluminescent-free TiO2 nanopores. ACS Nano 12, 11648–11656 (2018).
Cardozo, N. et al. Multiplexed direct detection of barcoded protein reporters on a nanopore array. Nat. Biotechnol. 40, 42–46 (2022).
Kennedy, E., Dong, Z., Tennant, C. & Timp, G. Reading the primary structure of a protein with 0.07-nm3 resolution using a subnanometre-diameter pore. Nat. Nanotechnol. 11, 968–976 (2016).
Rosen, C. B., Rodriguez-Larrea, D. & Bayley, H. Single-molecule site-specific detection of protein phosphorylation with a nanopore. Nat. Biotechnol. 32, 179–181 (2014).
Restrepo-Pérez, L., Wong, C. H., Maglia, G., Dekker, C. & Joo, C. Label-free detection of post-translational modifications with a nanopore. Nano Lett. 19, 7957–7964 (2019).
Behrends, J. C. Resolving isomeric posttranslational modifications using a biological nanopore as a sensor of molecular shape. J. Am. Chem. Soc. 144, 16060–16068 (2022).
Ohshiro, T. et al. Detection of post-translational modifications in single peptides using electron tunnelling currents. Nat. Nanotechnol. 9, 835–840 (2014).
Zhao, Y. et al. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat. Nanotechnol. 9, 466–473 (2014).
Brinkerhoff, D. H. & Dekker, C. Protein and peptide fingerprinting and sequencing by nanopore translocation of peptide-oligonucleotide complexes. PCT patent 2021/133168 A1 (2021).
Heron, J. A., Edward, G. J. & Slawa, S. M. Method of characterising a target polypeptide using a nanopore. PCT patent 2021/111125 A1 (2021).
Nova, I. C. et al. Detection of phosphorylation post-translational modifications along single peptides with nanopores. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01839-z (2023).
Boskovic, F. & Keyser, U. F. Toward single-molecule proteomics. Science 374, 1443–1444 (2021).
Wanunu, M. Back and forth with nanopore peptide sequencing. Nat. Biotechnol. 40, 172–173 (2022).
Aggarwal, V. & Ha, T. Single-molecule fluorescence microscopy of native macromolecular complexes. Curr. Opin. Struct. Biol. 41, 225–232 (2016).
Cohen, L. & Walt, D. R. Single-molecule arrays for protein and nucleic acid analysis. Annu. Rev. Anal. Chem. 10, 345–363 (2017).
Edman, P. A method for the determination of the amino acid sequence in peptides. Arch. Biochem. 22, 475–476 (1949).
Swaminathan, J. et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol. 36, 1076–1082 (2018).
Reed, B. D. et al. Real-time dynamic single-molecule protein sequencing on an integrated semiconductor device. Science 378, 186–192 (2022).
Kafader, J. O. et al. Measurement of individual ions sharply increases the resolution of Orbitrap mass spectra of proteins. Anal. Chem. 91, 2776–2783 (2019).
Makarov, A. & Denisov, E. Dynamics of ions of intact proteins in the Orbitrap mass analyzer. J. Am. Soc. Mass Spectrom. 20, 1486–1495 (2009).
Rose, R. J., Damoc, E., Denisov, E., Makarov, A. & Heck, A. J. R. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat. Methods 9, 1084–1086 (2012).
Kafader, J. O. et al. Multiplexed mass spectrometry of individual ions improves measurement of proteoforms and their complexes. Nat. Methods 17, 391–394 (2020).
Mock, A., Braun, M., Scholl, C., Fröhling, S. & Erkut, C. Transcriptome profiling for precision cancer medicine using shallow nanopore cDNA sequencing. Sci. Rep. 13, 2378 (2023).
Vermeulen, C. et al. Ultra-fast deep-learned CNS tumour classification during surgery. Nature 622, 842–849 (2023).
Bennett, H. M., Stephenson, W., Rose, C. M. & Darmanis, S. Single-cell proteomics enabled by next-generation sequencing or mass spectrometry. Nat. Methods 20, 363–374 (2023).
Motone, K. & Nivala, J. Not if but when nanopore protein sequencing meets single-cell proteomics. Nat. Methods 20, 336–338 (2023).
Matzinger, M., Müller, E., Dürnberger, G., Pichler, P. & Mechtler, K. Robust and easy-to-use one-pot workflow for label-free single-cell proteomics. Anal. Chem. 95, 4435–4445 (2023).
Candia, J. et al. Assessment of variability in the SOMAscan assay. Sci. Rep. 7, 14248 (2017).
Niu, M. et al. Droplet-based transcriptome profiling of individual synapses. Nat. Biotechnol. 41, 1332–1344 (2023).
Mamanova, L. & Turner, D. J. Low-bias, strand-specific transcriptome Illumina sequencing by on-flowcell reverse transcription (FRT-seq). Nat. Protoc. 6, 1736–1747 (2011).
Koch, C. et al. Nanopore sequencing of DNA-barcoded probes for highly multiplexed detection of microRNA, proteins and small biomarkers. Nat. Nanotechnol. 18, 1483–1491 (2023).
Liu, J. & Aksimentiev, A. Molecular determinants of current blockade produced by peptide transport through a nanopore. ACS Nanosci. https://doi.org/10.1021/acsnanoscienceau.3c00046 (2023).
Mahendran, K. R. et al. A monodisperse transmembrane α-helical peptide barrel. Nat. Chem. 9, 411–419 (2017).
Xu, C. et al. Computational design of transmembrane pores. Nature 585, 129–134 (2020).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996).
Caldwell, C. C. & Spies, M. Helicase SPRNTing through the nanopore. Proc. Natl Acad. Sci. USA 114, 11809–11811 (2017).
Mardis, E. R. Next-generation sequencing platforms. Annu. Rev. Anal. Chem. 6, 287–303 (2013).
Goodwin, S., McPherson, J. D. & McCombie, W. R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351 (2016).
Fuller, C. W. et al. The challenges of sequencing by synthesis. Nat. Biotechnol. 27, 1013–1023 (2009).
Kandaswamy, V., Eugene, T. & Bernard, M. A. Nucleic acid sequencing methods and systems. PCT patent 2017/014762 A1 (2017).
Meslier, V. et al. Benchmarking second and third-generation sequencing platforms for microbial metagenomics. Sci. Data 9, 694 (2022).
Füllgrabe, J. et al. Simultaneous sequencing of genetic and epigenetic bases in DNA. Nat. Biotechnol. 41, 1457–1464 (2023).
Rohs, R. et al. The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253 (2009).
Remaut, H., Jayasinghe, L. & Howorka, S. Mutant CsgG Pores. PCT patent 2016/034591 A3 (2016).
Iinuma, R. et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344, 65–69 (2014).
Schaus, T. E., Woo, S., Xuan, F., Chen, X. & Yin, P. A DNA nanoscope via auto-cycling proximity recording. Nat. Commun. 8, 696 (2017).
Filius, M., Kim, S. H., Severins, I. & Joo, C. High-resolution single-molecule FRET via DNA eXchange (FRET X). Nano Lett. 21, 3295–3301 (2021).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Obradovic, A. et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell 184, 2988–3005.e16 (2021).
Trzupek, D. et al. Discovery of CD80 and CD86 as recent activation markers on regulatory T cells by protein-RNA single-cell analysis. Genome Med. 12, 55 (2020).
Lundberg, M., Eriksson, A., Tran, B., Assarsson, E. & Fredriksson, S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 39, e102 (2011).
Frei, A. P. et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat. Methods 13, 269–275 (2016).
Acknowledgements
The Howorka Group receives funding from the Human Frontiers Science Program (RGP0047/2020), the Engineering and Physical Sciences Research Council (EP/N009282/1 and EP/V02874X/1), the Wellcome Institutional Strategic Support Fund, the Moorfields Biomedical Research Centre and Oxford Nanopore Technologies plc. We thank J. Ciccone, M. Booth, and K. F. Au for critically reading the manuscript, J. Ciccone and A. Rottensteiner for editing the figures, and J. Ciccone for helping edit the text.
Author information
Authors and Affiliations
Contributions
A.D. and S.H. contributed to discussions and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.H. is named inventor on a patent on the CsgG nanopore which is licensed to Oxford Nanopore Technologies Ltd. S.H. and A.D. are named inventors on patents on DNA nanopores which are licensed to Oxford Nanopore Technologies Ltd.
Peer review
Peer review information
Nature Chemistry thanks Abdelghani Oukhaled and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dorey, A., Howorka, S. Nanopore DNA sequencing technologies and their applications towards single-molecule proteomics. Nat. Chem. 16, 314–334 (2024). https://doi.org/10.1038/s41557-023-01322-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41557-023-01322-x