Abstract
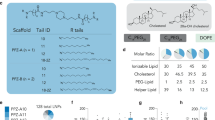
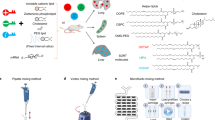
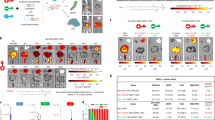
Endosomal escape remains a fundamental barrier hindering the advancement of nucleic acid therapeutics. Taking inspiration from natural phospholipids that comprise biological membranes, we report the combinatorial synthesis of multi-tailed ionizable phospholipids (iPhos) capable of delivering messenger RNA or mRNA/single-guide RNA for gene editing in vivo. Optimized iPhos lipids are composed of one pH-switchable zwitterion and three hydrophobic tails, which adopt a cone shape in endosomal acidic environments to facilitate membrane hexagonal transformation and subsequent cargo release from endosomes. Structure–activity relationships reveal that iPhos chemical structure can control in vivo efficacy and organ selectivity. iPhos lipids synergistically function with various helper lipids to formulate multi-component lipid nanoparticles (called iPLNPs) for selective organ targeting. Zwitterionic, ionizable cationic and permanently cationic helper lipids enable tissue-selective mRNA delivery and CRISPR–Cas9 gene editing in spleen, liver and lungs (respectively) following intravenous administration. This rational design of functional phospholipids demonstrates substantial value for gene editing research and therapeutic applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
All relevant data supporting the findings of this study are available within the paper and Supplementary Information. The raw data is available from the corresponding author upon request.
References
Wang, H. X. et al. CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem. Rev. 117, 9874–9906 (2017).
Hajj, K. A. & Whitehead, K. A. Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2, 17056 (2017).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Shahbazi, R. et al. Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nat. Mater. 18, 1124–1132 (2019).
Yin, H. et al. Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat. Biotechnol. 35, 1179 (2017).
Miller, J. B. et al. Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew. Chem. Int. Ed. 56, 1059–1063 (2017).
Liu, J. et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv. Mater. 31, 1902575 (2019).
Kanasty, R., Dorkin, J. R., Vegas, A. & Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 12, 967–977 (2013).
Semple, S. C. et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 28, 172–176 (2010).
Jayaraman, M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. 51, 8529–8533 (2012).
Love, K. et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl Acad. Sci. USA 107, 1864–1869 (2010).
Cheng, Q. et al. Dendrimer-based lipid nanoparticles deliver therapeutic FAH mRNA to normalize liver function and extend survival in a mouse model of hepatorenal tyrosinemia type I. Adv. Mater. 30, e1805308 (2018).
Sabnis, S. et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 26, 1509–1519 (2018).
van Meer, G., Voelker, D. R. & Feigenson, G. W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 (2008).
Miller, J. B., Kos, P., Tieu, V., Zhou, K. & Siegwart, D. J. Development of cationic quaternary ammonium sulfonamide amino lipids for nucleic acid delivery. ACS Appl. Mater. Inter. 10, 2302–2311 (2018).
Gilleron, J. et al. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 31, 638–646 (2013).
Wittrup, A. et al. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 33, 870–876 (2015).
Miller, J. B. & Siegwart, D. J. Design of synthetic materials for intracellular delivery of RNAs: from siRNA-mediated gene silencing to CRISPR/Cas gene editing. Nano Res. 11, 5310–5337 (2018).
Whitehead, K. A. et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 5, 4277 (2014).
Miao, L. et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 37, 1174–1185 (2019).
Kaczmarek, J. C. et al. Optimization of a degradable polymer–lipid nanoparticle for potent systemic delivery of mRNA to the lung endothelium and immune cells. Nano Lett. 18, 6449–6454 (2018).
Paunovska, K. et al. Nanoparticles containing oxidized cholesterol deliver mRNA to the liver microenvironment at clinically relevant doses. Adv. Mater. 31, 1807748 (2019).
Fenton, O. S. et al. Bioinspired alkenyl amino alcohol ionizable lipid materials for highly potent in vivo mRNA delivery. Adv. Mater. 28, 2939–2943 (2016).
Truong, B. et al. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc. Natl Acad. Sci. USA 116, 21150–21159 (2019).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue specific mRNA delivery and CRISPR/Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Hajj, K. A. et al. Branched-tail lipid nanoparticles potently deliver mRNA In vivo due to enhanced ionization at endosomal pH. Small 15, 1805097 (2019).
Fenton, O. S. et al. Synthesis and biological evaluation of ionizable lipid materials for the in vivo delivery of messenger RNA to B lymphocytes. Adv. Mater. 29, 1606944 (2017).
Alabi, C. A. et al. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc. Natl Acad. Sci. USA 110, 12881–12886 (2013).
Hou, X. et al. Vitamin lipid nanoparticles enable adoptive macrophage transfer for the treatment of multidrug-resistant bacterial sepsis. Nat. Nanotechnol. 15, 41–46 (2020).
Menger, F. M. & Peresypkin, A. V. A combinatorially-derived structural phase diagram for 42 zwitterionic geminis. J. Am. Chem. Soc. 123, 5614–5615 (2001).
Wang, D. et al. Supramolecularly engineered phospholipids constructed by nucleobase molecular recognition: upgraded generation of phospholipids for drug delivery. Chem. Sci. 6, 3775–3787 (2015).
Akinc, A. et al. A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol. 26, 561–569 (2008).
Zhou, K. et al. Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model. Proc. Natl Acad. Sci. USA 113, 520–525 (2016).
Liu, S. et al. Highly branched poly(β-amino ester) delivery of minicircle DNA for transfection of neurodegenerative disease related cells. Nat. Commun. 10, 3307 (2019).
Zhou, J. et al. Biodegradable poly(amine-co-ester) terpolymers for targeted gene delivery. Nat. Mater. 11, 82–90 (2012).
Liu, S. et al. Bioreducible zinc(II)-coordinative polyethylenimine with low molecular weight for robust gene delivery of primary and stem cells. J. Am. Chem. Soc. 139, 5102–5109 (2017).
Dahlman, J. E. et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 9, 648–655 (2014).
Schlame, M. et al. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nat. Chem. Bio. 8, 862–869 (2012).
Hafez, I. M., Maurer, N. & Cullis, P. R. On the mechanism whereby cationic lipids promote intracellular delivery of polynucleic acids. Gene Ther. 8, 1188–1196 (2001).
Wei, T. et al. Anticancer drug nanomicelles formed by self-assembling amphiphilic dendrimer to combat cancer drug resistance. Proc. Natl Acad. Sci. USA 112, 2978–2983 (2015).
Walsh, C. L., Nguyen, J. & Szoka, F. C. Synthesis and characterization of novel zwitterionic lipids with pH-responsive biophysical properties. Chem. Commun. 48, 5575–5577 (2012).
Zhang, Y. et al. The development of an in vitro assay to screen lipid based nanoparticles for siRNA delivery. J. Controlled Release 174, 7–14 (2014).
Cheng, Y., Yumul, R. C. & Pun, S. H. Virus-inspired polymer for efficient in vitro and in vivo gene delivery. Angew. Chem. Int. Ed. 55, 12013–12017 (2016).
Zhou, D. et al. The transition from linear to highly branched poly(beta-amino ester)s: branching matters for gene delivery. Sci. Adv. 2, e1600102 (2016).
Kowalski, P. S. et al. Ionizable amino‐polyesters synthesized via ring opening polymerization of tertiary amino‐alcohols for tissue selective mRNA delivery. Adv. Mater. 30, 1801151 (2018).
Li, B. et al. An orthogonal array optimization of lipid-like nanoparticles for mRNA delivery in vivo. Nano Lett. 15, 8099–8107 (2015).
Wei, T., Cheng, Q., Min, Y. L., Olson, E. N. & Siegwart, D. J. Systemic nanoparticle delivery of CRISPR-Cas9 ribonucleoproteins for effective tissue specific genome editing. Nat. Commun. 11, 3232 (2020).
Akinc, A. et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 14, 1084–1087 (2019).
Patel, S. et al. Naturally-occurring cholesterol analogues in lipid nanoparticles induce polymorphic shape and enhance intracellular delivery of mRNA. Nat. Commun. 11, 983 (2020).
Xue, W. et al. CRISPR-mediated direct mutation of cancer genes in the mouse liver. Nature 514, 380–384 (2014).
Staahl, B. T. et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 35, 431–434 (2017).
Acknowledgements
D.J.S. acknowledges financial support from the National Institutes of Health (NIH) National Institute of Biomedical Imaging and Bioengineering (NIBIB) (grant no. R01 EB025192-01A1), the American Cancer Society (ACS) (grant no. RSG-17-012-01), the Welch Foundation (grant no. I-1855) and the Cystic Fibrosis Foundation (CFF) (grant no. SIEGWA18XX0). T.W. acknowledges financial support from the Cancer Prevention and Research Institute of Texas (CPRIT) Training grant (no. RP160157). We acknowledge the UTSW Tissue Resource, supported in part by the National Cancer Institute (grant no. 5P30CA142543) and the Moody Foundation Flow Cytometry Facility.
Author information
Authors and Affiliations
Contributions
S.L., Q.C. and D.J.S. designed the research. S.L., Q.C., T.W., X.Y., L.T.J. and L.F. performed the experiments. All the authors were involved in the data analyses. S.L. and D.J.S. wrote the manuscript, and all authors discussed and commented on it. D.J.S. directed the research.
Corresponding author
Ethics declarations
Competing interests
D.J.S., S.L., Q.C., T.W. and X.Y., and the Reagents of the University of Texas System have filed a patent application on this technology.
Additional information
Peer review information Nature Materials thanks Bruno Pitard, John Rossi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–32, Tables 1–3 and Methods.
Rights and permissions
About this article
Cite this article
Liu, S., Cheng, Q., Wei, T. et al. Membrane-destabilizing ionizable phospholipids for organ-selective mRNA delivery and CRISPR–Cas gene editing. Nat. Mater. 20, 701–710 (2021). https://doi.org/10.1038/s41563-020-00886-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41563-020-00886-0
This article is cited by
-
Strategies for non-viral vectors targeting organs beyond the liver
Nature Nanotechnology (2024)
-
Designer phospholipid capping ligands for soft metal halide nanocrystals
Nature (2024)
-
Liquid crystalline inverted lipid phases encapsulating siRNA enhance lipid nanoparticle mediated transfection
Nature Communications (2024)
-
Combinatorial development of nebulized mRNA delivery formulations for the lungs
Nature Nanotechnology (2024)
-
Natural long-chain saturated fatty acids doped LNPs enabling spleen selective mRNA translation and potent cancer immunotherapy
Nano Research (2024)