Abstract
Despite the well-established role of nuclear organization in the regulation of gene expression, little is known about the reverse: how transcription shapes the spatial organization of the genome. Owing to the small sizes of most previously studied genes and the limited resolution of microscopy, the structure and spatial arrangement of a single transcribed gene are still poorly understood. Here we study several long highly expressed genes and demonstrate that they form open-ended transcription loops with polymerases moving along the loops and carrying nascent RNAs. Transcription loops can span across micrometres, resembling lampbrush loops and polytene puffs. The extension and shape of transcription loops suggest their intrinsic stiffness, which we attribute to decoration with multiple voluminous nascent ribonucleoproteins. Our data contradict the model of transcription factories and suggest that although microscopically resolvable transcription loops are specific for long highly expressed genes, the mechanisms underlying their formation could represent a general aspect of eukaryotic transcription.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Hi-C data have been uploaded to Gene Expression Omnibus (GEO) and are available under accession GSE150704. ChIP–seq and RNA-seq data are available at ArrayExpress (EMBL-EBI) under accession E-MTAB-9060 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-9060/). Previously published reference genome mm10 and gene annotation of the C57BL/6 J strain were downloaded from the Ensemble database (version GRCm38, release 74). Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Code availability
The used code for the measurement of flank distances is available at https://github.com/hoerldavid/fish_analysis; code used for the analysis of Hi-C data (Cooler, Cooltools, Distiller, Pairtools) is available at https://github.com/open2c; the code used for polymer simulations is available at https://github.com/mirnylab/openmm-polymer-legacy.
References
Andersson, R. & Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 21, 71–87 (2020).
Cramer, P. Organization and regulation of gene transcription. Nature 573, 45–54 (2019).
Herzel, L., Straube, K. & Neugebauer, K. M. Long-read sequencing of nascent RNA reveals coupling among RNA processing events. Genome Res. 28, 1008–1019 (2018).
Feodorova, Y., Falk, M., Mirny, L. A. & Solovei, I. Viewing nuclear architecture through the eyes of nocturnal mammals. Trends Cell Biol. 30, 276–289 (2020).
Solovei, I., Thanisch, K. & Feodorova, Y. How to rule the nucleus: divide et impera. Curr. Opin. Cell Biol. 40, 47–59 (2016).
van Steensel, B. & Belmont, A. S. Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017).
Macgregor, H. C. An Introduction to Animal Cytogenetics (Chapman & Hall, 1993).
Bjork, P. & Wieslander, L. The Balbiani ring story: synthesis, assembly, processing, and transport of specific messenger RNA–protein complexes. Annu. Rev. Biochem. 84, 65–92 (2015).
Chakalova, L. & Fraser, P. Organization of transcription. Cold Spring Harb. Perspect. Biol. 2, a000729 (2010).
Osborne, C. S. et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36, 1065–1071 (2004).
Schoenfelder, S. et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42, 53–61 (2010).
Cook, P. R. The organization of replication and transcription. Science 284, 1790–1795 (1999).
Papantonis, A. & Cook, P. R. Transcription factories: genome organization and gene regulation. Chem. Rev. 113, 8683–8705 (2013).
Mateo, L. J. et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Rodermund, L. et al. Time-resolved structured illumination microscopy reveals key principles of Xist RNA spreading. Science 372, (2021).
Morgan, G. T. Imaging the dynamics of transcription loops in living chromosomes. Chromosoma 127, 361–374 (2018).
Schermelleh, L., Heintzmann, R. & Leonhardt, H. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190, 165–175 (2010).
Jjingo, D., Huda, A., Gundapuneni, M., Marino-Ramirez, L. & Jordan, I. K. Effect of the transposable element environment of human genes on gene length and expression. Genome Biol. Evol. 3, 259–271 (2011).
Hutchison, N. Lampbrush chromosomes of the chicken, Gallus domesticus. J. Cell Biol. 105, 1493–1500 (1987).
Miller, O. L. & Beatty, B. R. Visualization of nuclear genes. Science 164, 955–957 (1969).
Kaufmann, R., Cremer, C. & Gall, J. G. Superresolution imaging of transcription units on newt lampbrush chromosomes. Chromosome Res. 20, 1009–1015 (2012).
Carmo-Fonseca, M. & Kirchhausen, T. The timing of pre-mRNA splicing visualized in real-time. Nucleus 5, 11–14 (2014).
Ansari, A. & Hampsey, M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev. 19, 2969–2978 (2005).
Larsson, A. J. M. et al. Genomic encoding of transcriptional burst kinetics. Nature 565, 251–254 (2019).
Tantale, K. et al. A single-molecule view of transcription reveals convoys of RNA polymerases and multi-scale bursting. Nat. Commun. 7, 12248 (2016).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Bensaude, O. Inhibiting eukaryotic transcription: which compound to choose? How to evaluate its activity? Transcription 2, 103–108 (2011).
Mahy, N. L., Perry, P. E., Gilchrist, S., Baldock, R. A. & Bickmore, W. A. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J. Cell Biol. 157, 579–589 (2002).
Brown, J. M. et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J. Cell Biol. 182, 1083–1097 (2008).
Chambeyron, S., Da Silva, N. R., Lawson, K. A. & Bickmore, W. A. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132, 2215–2223 (2005).
Mahy, N. L., Perry, P. E. & Bickmore, W. A. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159, 753–763 (2002).
Volpi, E. V. et al. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113, 1565–1576 (2000).
Williams, R. R., Broad, S., Sheer, D. & Ragoussis, J. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell. Res. 272, 163–175 (2002).
Abramo, K. et al. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat. Cell Biol. 21, 1393–1402 (2019).
Kalhor, R., Tjong, H., Jayathilaka, N., Alber, F. & Chen, L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat. Biotechnol. 30, 90–98 (2012).
Shah, S. et al. Dynamics and spatial genomics of the nascent transcriptome by intron seqFISH. Cell 174, 363–376 e316 (2018).
Hsieh, T. S. et al. Resolving the 3D landscape of transcription-linked mammalian chromatin folding. Mol. Cell 78, 539–553 (2020).
Banigan, E. J. & Mirny, L. A. The interplay between asymmetric and symmetric DNA loop extrusion. eLife 9, (2020).
Brandao, H. B. et al. RNA polymerases as moving barriers to condensin loop extrusion. Proc. Natl Acad. Sci. USA 116, 20489–20499 (2019).
Banigan, E. J. et al. Transcription shapes 3D chromatin organization by interacting with loop-extruding cohesin complexes. https://doi.org/10.1101/2022.01.07.475367 (2022).
Muller-McNicoll, M. & Neugebauer, K. M. How cells get the message: dynamic assembly and function of mRNA-protein complexes. Nat. Rev. Genet. 14, 275–287 (2013).
Olins, A. L. & Olins, D. E. Spheroid chromatin units (v bodies). Science 183, 330–332 (1974).
Liu, X., Farnung, L., Wigge, C. & Cramer, P. Cryo-EM structure of a mammalian RNA polymerase II elongation complex inhibited by alpha-amanitin. J. Biol. Chem. 293, 7189–7194 (2018).
Paturej, J., Sheiko, S. S., Panyukov, S. & Rubinstein, M. Molecular structure of bottlebrush polymers in melts. Sci. Adv. 2, e1601478 (2016).
Kotake, Y. et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3, 570–575 (2007).
Mirny, L. A. & Solovei, I. Keeping chromatin in the loop(s). Nat. Rev. Mol. Cell Biol. 22, 439–440 (2021).
Cho, W. K. et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361, 412–415 (2018).
Guo, Y. E. et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 572, 543–548 (2019).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. A phase separation model for transcriptional control. Cell 169, 13–23 (2017).
Cisse, I. I. et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341, 664–667 (2013).
Henninger, J. E. et al. RNA-mediated feedback control of transcriptional condensates. Cell 184, 207–225 e224 (2021).
Hampsey, M., Singh, B. N., Ansari, A., Laine, J. P. & Krishnamurthy, S. Control of eukaryotic gene expression: gene loops and transcriptional memory. Adv. Enzyme Regul. 51, 118–125 (2011).
Martin, M., Cho, J., Cesare, A. J., Griffith, J. D. & Attardi, G. Termination factor-mediated DNA loop between termination and initiation sites drives mitochondrial rRNA synthesis. Cell 123, 1227–1240 (2005).
Singh, B. N. & Hampsey, M. A transcription-independent role for TFIIB in gene looping. Mol. Cell 27, 806–816 (2007).
Lee, K., Hsiung, C. C., Huang, P., Raj, A. & Blobel, G. A. Dynamic enhancer–gene body contacts during transcription elongation. Genes Dev. 29, 1992–1997 (2015).
Zheng, M. et al. Multiplex chromatin interactions with single-molecule precision. Nature 566, 558–562 (2019).
Cremer, T. & Cremer, M. Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010).
Keizer, V. I. P. et al. Live-cell micromanipulation of a genomic locus reveals interphase chromatin mechanics. Preprint at bioRxiv https://doi.org/10.1101/2021.04.20.439763 (2021).
Khanna, N., Zhang, Y., Lucas, J. S., Dudko, O. K. & Murre, C. Chromosome dynamics near the sol-gel phase transition dictate the timing of remote genomic interactions. Nat. Commun. 10, 2771 (2019).
Strickfaden, H. et al. Condensed chromatin behaves like a solid on the mesoscale in vitro and in living cells. Cell 183, 1772–1784 e1713 (2020).
Bagnoli, J. W. et al. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun. 9, 2937 (2018).
Ziegenhain, C. et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643 e634 (2017).
Parekh, S., Ziegenhain, C., Vieth, B., Enard, W. & Hellmann, I. zUMIs—a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience 7, (2018).
Rau, A., Gallopin, M., Celeux, G. & Jaffrezic, F. Data-based filtering for replicated high-throughput transcriptome sequencing experiments. Bioinformatics 29, 2146–2152 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Link, S. et al. PWWP2A binds distinct chromatin moieties and interacts with an MTA1-specific core NuRD complex. Nat. Commun. 9, 4300 (2018).
Punzeler, S. et al. Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation. EMBO J. 36, 2263–2279 (2017).
Belaghzal, H., Dekker, J. & Gibcus, J. H. Hi-C 2.0: an optimized Hi-C procedure for high-resolution genome-wide mapping of chromosome conformation. Methods 123, 56–65 (2017).
Abdennur, N. & Mirny, L. A. Cooler: scalable storage for Hi-C data and other genomically labeled arrays. Bioinformatics 36, 311–316 (2020).
Imakaev, M. et al. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003 (2012).
Cremer, M. et al. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol. Biol. 463, 205–239 (2008).
Kishi, J. Y. et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods 16, 533–544 (2019).
Kishi, J. Y., Schaus, T. E., Gopalkrishnan, N., Xuan, F. & Yin, P. Programmable autonomous synthesis of single-stranded DNA. Nat. Chem. 10, 155–164 (2018).
Bienko, M. et al. A versatile genome-scale PCR-based pipeline for high-definition DNA FISH. Nat. Methods 10, 122–124 (2013).
Solovei, I. & Cremer, M. 3D-FISH on cultured cells combined with immunostaining. Methods Mol. Biol. 659, 117–126 (2010).
Solovei, I. et al. in FISH: A Practical Approach (eds Beatty, B. et al.) 119–157 (Oxford Univ. Press, 2002).
Solovei, I. Fluorescence in situ hybridization (FISH) on tissue cryosections. Methods Mol. Biol. 659, 71–82 (2010).
Eberhart, A. et al. Epigenetics of eu- and heterochromatin in inverted and conventional nuclei from mouse retina. Chromosome Res. 21, 535–554 (2013).
Eberhart, A., Kimura, H., Leonhardt, H., Joffe, B. & Solovei, I. Reliable detection of epigenetic histone marks and nuclear proteins in tissue cryosections. Chromosome Res. 20, 849–858 (2012).
Solovei, I. et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013).
Walter, J. et al. Towards many colors in FISH on 3D-preserved interphase nuclei. Cytogenet Genome Res. 114, 367–378 (2006).
Bystricky, K., Heun, P., Gehlen, L., Langowski, J. & Gasser, S. M. Long-range compaction and flexibility of interphase chromatin in budding yeast analyzed by high-resolution imaging techniques. Proc. Natl Acad. Sci. USA 101, 16495–16500 (2004).
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Rippe, K. Making contacts on a nucleic acid polymer. Trends Biochem. Sci 26, 733–740 (2001).
Ou, H. D. et al. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357, (2017).
Hajjoul, H. et al. High-throughput chromatin motion tracking in living yeast reveals the flexibility of the fiber throughout the genome. Genome Res. 23, 1829–1838 (2013).
Lucas, J. S., Zhang, Y., Dudko, O. K. & Murre, C. 3D trajectories adopted by coding and regulatory DNA elements: first-passage times for genomic interactions. Cell 158, 339–352 (2014).
Nuebler, J., Fudenberg, G., Imakaev, M., Abdennur, N. & Mirny, L. A. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl Acad. Sci. USA 115, E6697–E6706 (2018).
Acknowledgements
We are grateful to D. Hörl and J. Ryan for the help with ImageJ plugins and programming. We thank A. Maiser and K. Brandstetter for the help with high-resolution microscopy. We acknowledge J. Bates, D. Eick, P. Becker, M. Carmo-Fonseca, A. Olins and D. Olins and H.C.Macgregor for fruitful and insightful discussions. We thank D. Kralev and T. Suzuki for the help with animation. This work has been supported by the Deutsche Forschungsgemeinschaft grants (SO1054/1 and SP2202/SO1054/2 to I.S., SPP 2202/LE721/17-1 to H.L. and SFB1064 to I.S. and H.L.) and National Institutes of Health grants (HG007743 to H.L., HG003143 to J.D. and GM114190 to L.M. by the Center for 3D Structure and Physics of the Genome of NIH 4DN Consortium, DK107980). J.D. is an investigator of the Howard Hughes Medical Institute. I.S. is deeply thankful to H. C. Macgregor for his guidance.
Author information
Authors and Affiliations
Contributions
I.S. conceived the project. S. Leidescher, S.U., Y.F., K.T. and I.S. obtained biological samples. I.S., Y.F., S. Leidescher and S.U. conceived and performed microscopy and image analysis. S. Leidescher, C.M. and S. Link performed RNA-seq and ChIP–seq experiments. S.B. performed RNA-seq and ChIP–seq analyses. Y.F. and E.H. performed Hi-C experiments. E.H., J.D., J.R. (formerly known as J. Nübler), A.G. and L.M. performed Hi-C analysis. J.R. with contribution from L.M. performed simulations. S. Leidescher, S.U., J.R., A.G. and I.S. prepared the figures. I.S. wrote the manuscript with contributions from S. Leidescher, Y.F., S.U., J.R., K.T., E.H., H.L., J.D. and L.M.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Long genes are rare and expressed at lower levels than short genes.
a, Analysis of gene length distribution within the human and mouse genomes showed that about 43% and 46% of all protein coding genes, respectively, have a length ≤20 kb and only 18% and 14% have a length of 100 kb or above. Bin size: 20 kb. Genes are annotated according to GENCODE. Only genes with a length <500 kb are shown. b, To select suitable genes for visualization with light microscopy, we studied gene expression profiles across 50 human tissues using the publicly available Genotype-Tissue Expression database (GTEx Consortium) and found that long genes, as a rule, are not highly expressed. For example, in liver (top) and brain (bottom) there were no expressed genes with both a length ≥100 kb and with a median expression ≥1,000 TPM. c, Comparison of RNAPII occupancy between short and long expressed genes. ChIP-seq with an antibody against the CTD of RNAPII in cultured mouse myoblasts (left) and in vitro differentiated myotubes (right). All genes, expressed (>1 TPM, blue) and silent (<1 TPM, red), were split into five categories according to their size. RNAPII density (Y-axis) is plotted against the respective position within the gene (X-axis); each gene is divided into 200 equally sized bins and genes from the same size category are aligned according to the bins. Expressed genes display a higher occupancy with RNAPII compared to non-expressed genes, especially in the TSS region. In the group of expressed genes, the RNAPII occupancy negatively correlates with gene length: the shorter the genes, the higher the RNAPII occupancy. d, Analysis of RNA-seq data for myoblasts (left) and myotubes (right). The median expression level (TPM) is higher in groups containing shorter genes (<25 kb) and generally negatively correlates with gene length.
Extended Data Fig. 2 Visualization of the five selected genes in expressing and not expressing cells.
a, The Tg gene is expressed in thyrocytes where both alleles form prominent TLs expanding into the nuclear interior. In neighboring cells with a silent Tg gene - parathyroid gland cells, tracheal chondrocytes, epithelial cells, fibroblasts and muscles - Tg is highly condensed and sequestered to the nuclear periphery. b, The Ttn gene is expressed in skeletal muscle (b1), heart muscle (b2) and myotubes differentiated from Pmi28 myoblasts in vitro (b3). Note that only muscle nuclei (solid arrowheads) exhibit TLs. In muscle fibroblasts (arrows) or undifferentiated cultured myoblasts (empty arrowheads), Ttn is condensed at the nuclear periphery. c, The Neb gene is expressed in skeletal muscles and cultured myotubes, although to a lesser degree than Ttn. Accordingly, it forms smaller TLs. Arrowheads indicate muscle nuclei; arrows indicate fibroblast nuclei with silent Neb. d, e, The Myh11 (d) and Cald1 (e) genes are expressed in smooth muscles of colon and bladder where they form TLs. Note that after RNA-FISH, only smooth muscles (arrowheads) but not the neighboring epithelial cells (arrows) exhibit TLs. In addition, Cald1 is expressed in cultured myoblasts and forms small TLs in these cells. As indicated above the panels, images display signals after either RNA-FISH (no tissue/cell DNA denaturation and no RNasing), or simultaneous detection of DNA and RNA (tissue/cell DNA denaturation but no RNasing). All images are projections of 1–3 µm confocal stacks. Scale bars for overviews of skeletal muscle, colon and bladder, 50 µm; for the rest of the panels, 5 µm. Data represent 100 in a,b3 and 10 in b1,b2,c-e independent experiments.
Extended Data Fig. 3 Structure and compaction of TLs.
a, The internal structure of Tg and Ttn TLs is not resolvable after deconvolution (left) and high resolution microscopy (mid, right). b, Coiling and folding of TLs demonstrated in 50–70 nm thin resin sections. The upper panel shows thin sections through nuclei of thyrocytes stained with DAPI (red) and Tg TLs detected by RNA-FISH (green). The lower panel shows 2-fold close-ups of the corresponding Tg TL as grey-scale images. Note curling and twisting of the loops. Images are single optical sections. c, To assess the compaction level of TLs, the contour length of three Tg TL regions was measured on projections after RNA-FISH. The track of the Segmented Line tool in ImageJ, used for measurements, is shown on the right panel. Tg regions of 153 kb, 109 kb and 62 kb had a similar compaction level and measured 9 µm, 6 µm and 4 µm, respectively. These values correspond to a nucleosomal structure of chromatin (table on the right). However, since Tg TLs display internal structures and since the measurements were performed on maximum intensity projections, the compaction level of Tg TLs is probably overestimated. Scale bars: a, 2 µm; b,c, 1 µm. Data represent 3 independent experiments in a-c.
Extended Data Fig. 4 TLs manifest co-transcriptional splicing.
Two sequentially positioned introns were labeled with oligoprobes encompassing 1.2–5 kb. The schematics above the panels depict the distribution of oligoprobes (green and red rectangles), labeled introns (green and red lines) and positions of BAC probes used as references (grey lines above genes). After RNA-FISH the intron probes label TLs only partially and sequentially. Since the 5’ and 3’ intron signals do not overlap, the 5’ introns are spliced before the 3’ introns are read. For instance, in Cald1, introns 1 and 3 are separated by intron 2, suggesting that the “green” intron 1 is spliced out before polymerases reach the “red” intron 3, most likely after RNAPII runs over the 3’ splice-site of the first intron. Projections of confocal sections through 2–3.5 µm. Scale bars: 2 µm, in close-ups, 1 µm. Data represent 2 independent experiments.
Extended Data Fig. 5 Close association of TLs with splicing factors.
Splicing factors (SON) and components of exon-junction complexes EIF4A3 and RBM8A (Y14), are either co-stained with RNAPII Ser2P (the two top rows) or visualized together with TLs in immuno-FISH (the rest of the rows). Note that signals of TLs and splicing factors colocalize only partly. The myotube nucleus is tetraploid and thus exhibits 4 Ttn RNA signals. Images are partial projections of either 0.6 µm (for immunostaining) or 0.9 µm (for immuno-FISH). Scale bars: 2 µm, in close-ups, 1 µm. Data represent 3 independent experiments.
Extended Data Fig. 6 Nucleoplasmic granules in cells with highly expressed long genes.
a, RNA-FISH reveals numerous nucleoplasmic granules (arrows) surrounding TLs (arrowheads) after hybridization with genomic probes. For clarity, only RNA signals are shown within the outlined nuclei. Empty arrowheads point at similar granules in the myotube cytoplasm. The asterisk marks the nucleus of a myoblast not expressing Ttn. b, In muscles and cultured myotubes, the majority of granules (81%) are double-labeled with probes for the 5’ and 3’ halves of Ttn and found in both the nucleoplasm and cytoplasm (arrows on the lower panel), thus likely representing Ttn mRNAs. Remarkably, the 5’ and 3’ signals are spatially distinguished within the granules (insertion) presumably due to the exceptionally long Ttn mRNA of ca 102 kb. The observed separation of the 5’ and 3’ halves of Ttn mRNA is in agreement with previously described structures of cytoplasmic mRNPs28,29,30. c, In difference to Ttn, mRNAs of Tg, Neb, Cald1 and Myh11 genes are short (4–20 kb) and can be only detected with oligoprobes specifically hybridizing to all exons. Thus the oligoprobe for all 48 Tg exons hybridizes to nRNAs decorating TLs (arrows) and also labels multiple nucleoplasmic granules (arrowheads). d, The majority of the other thyrocyte nucleoplasmic granules are labeled with either 5’ (green) or 3’ (red) genomic probes with only 10% of granules being double-labeled. The brightness of the RNA-signal on the most right panel is purposely increased to highlight nucleoplasmic granules (green and red arrows). Such differential labeling of nucleoplasmic granules, exemplified here for thyrocytes, is characteristic for other studied genes and strongly suggests that these granules represent accumulations of excised introns. The distribution of the used BAC probes in respect to the studied genes are depicted above the image panels. Scale bars: a, 2 µm for Tg and Myh11, 5 µm for Ttn and Neb; b, c, d, 2 µm. Data represent 10 in a,b and 3 in c,d independent experiments.
Extended Data Fig. 7 Fragments of Hi-C maps around TL-forming genes in corresponding tissues and cells.
Each row shows the same region around indicated genes in different tissues or cells; “on” indicates a cell type where a gene is active, allowing to compare changes associated with TL formation. Blue arrows indicate loss of TAD borders and associated dots of CTCF-CTCF enrichment for expressed Tg and Myh11. Red arrows indicate an increase in self-interactions within genes visible for expressed Ttn, Neb and Myh11.
Extended Data Fig. 8 Cis-to-trans contact ratios and A/B compartments in studied cells.
a, Cis-to-trans ratios and compartment affiliations for 5 studied genes. The scatter plots are computed from the compartment profiles and the cis-to-trans ratio profiles of the chromosomes harboring the genes at a bin size of 32 kb. Genes of interest are highlighted with red dots; the white crosses mark the chromosome means of the compartment and cis-to-trans ratio profiles. Tg, Ttn, Neb and Myh11 move from B to A compartment upon their activation; Cald1 is found in A compartment not only in myoblasts and smooth muscle, as expected, but also in thyroid and myotubes. This is in agreement with the low Cald1 expression also in thyroid samples enriched in blood vessels and in samples of myotubes that normally include up to 20% of myoblasts, as well as with localization of Cald1 in a gene-dense region of chromosome 6. Note that the Ttn and Neb genes tend to be in A compartment, although to a lesser degree, also in myoblasts, which can be explained by the presence of cells that started their differentiation into single-cell myotubes. b, Cis-to-trans ratios are lower in A than in B compartments for all four studied cell types. The scatter plots are computed from the genome wide compartment profile and the cis-to-trans ratio profile, both at a bin size of 1,024 kb. The Pearson correlation coefficients are indicated in the upper right corners of the scatter plots. c, Externalization of the expressed genes from their harboring chromosomes measured by the cis-to-trans ratios as a function of gene length and expression in corresponding tissues. Left column of heatmaps: median cis-to-trans normalized by chromosome. Notice the reduction of cis-to-trans (that is, increasing externalization) for highly transcribed genes (top rows in each heatmap) as gene length increases (moving from left to right). Similarly, cis-to-trans goes down for long genes (right-most column of each heatmap) as expression increases (going up along this column). Interestingly, lowly transcribed long genes (lower right corner) have high cis-to-trans, indicating strong internalization, but become strongly externalized as they become highly expressed (upper right corner). Middle column: median cis-to-trans ratios controlled for compartmental signal (the first eigenvector, E1). Heatmaps show the logarithm of observed median cis-to-trans ratio divided by the expected given E1 in the corresponding bin. For the expected value, all the genomic bins were separated into 20 ranges by their E1, and the median cis-to-trans for each range was considered as expected. Notice that for most genes, E1 explains most of the cis-to-trans ratio. However, cis-to-trans is considerably lower for extremely long and extremely highly transcribed genes (upper right corner of each heatmap). Right column: the number of genomic bins in each range of length and transcription. Notice that very few genes show high externalization. d, Table of coefficients of determination (R2) for regression of cis-to-trans ratios of the genomic bins normalized by chromosome, in four tissues. Only the bins of expressed genes (TPM > 100) are considered. Gene length is an excellent predictor of cis-to-trans ratio for genomic bins of highly expressed genes (TPM > 1000, 1st row), but not for other expressed genes (TPM from 100 to 1000, 2nd row). Gene expression is a good predictor of cis-to-trans ratio for genomic bins of long genes (TPM > 50 kb, 3rd row), but not for shorter genes (TPM < 50 kb, last row). Heatmaps: visual illustration of gene subsets in this analysis.
Extended Data Fig. 9 TLs do not cause insulation at different length scales.
Insulation assesses Hi-C contacts spanning across a given locus up to a maximal distance w (top right insert). Contacts in a square window of size w were aggregated and the square was slid along the Hi-C diagonal. The score was normalized by its genome wide mean. Profiles show log2 of the score, such that a locus with profile value -1 has a two-fold reduced number of contacts spanning the locus up to distance w compared to the genome wide mean. Insulation scores are computed with the cooltools package (https://github.com/mirnylab/cooltools). We computed insulation profiles for Hi-C maps with a bin size of 128 kb for various window sizes from 256 kb up to ≈16 Mb. For every analyzed gene, the left and right columns show a 3 and 20 Mb Hi-C map with insulation profiles for different window sizes; the top and bottom panels show insulation profiles in expressing (on) and non-expressing (off) cells, respectively. The analysis shows little correlation between insulation and the formation of TLs: insulation profiles at the gene loci do not differ much between cell types with the gene on or off. For example, the Tg gene shows a moderate dip at scales up to ≈ 1 Mb in both thyroids (on) and myoblasts (off), and no dip in either cell type on the larger scale. Analysis of simulated TLs (Fig. 8 and Extended data Fig. 10) confirmed that TL formation does not cause large scale insulation (bottom row).
Extended Data Fig. 10 Polymer simulation of chromosomes.
a, Six chromosomes (50 Mb each) were initiated in a mitotic-like state with unit volume density. Row 1 and 3 show top views, row 2 and 4 show side views. In rows 1 and 2 six chromosomes are differentially colored; in rows 3 and 4 compartmental segments of A and B type chromatin are differentially colored with red for A and blue for B compartments. The initial expansion is very fast (column 2). However, once the chromosomes fill the nucleus uniformly, the subsequent dynamics is very slow and chromosomes retain their territoriality (note that times increase logarithmically). Nevertheless, due to attraction of B-type chromatin to the lamina, a radial structure emerges (rows 3 and 4). b, TLs are modeled by choosing a 300 kb segment on each chromosome 25.4 minutes after expansion and increasing the stiffness of the polymer fiber. The genes quickly expand on the order of minutes and are simulated for approximately 1.5 h. The measurements of inter-flank distances and Hi-C maps are performed using configurations sampled from the second half of this time interval. When genes are deactivated by removing the excess stiffness, they collapse back to the inactive state. c, Left: Hi-C of all 6 chromosomes shows their territoriality as patches. Second-left: A Hi-C contact map averaged over all 6 chromosomes exhibits the checkerboard pattern of a typical segregation of A- and B-type chromatin. The three rightmost graphs show zoomed views of modeled genes with stiffness profiles above the maps.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5.
Supplementary Table 1
Excel spreadsheet includes the distribution of human genes according to their lengths, a list of protein-coding genes and their expression levels in different human tissues, and a list of selected genes with lengths larger than 100 kb and expression levels of ca. 1,000 TPM.
Supplementary Table 2
Excel spreadsheet includes the distribution of mouse genes according to their lengths, a list of protein-coding genes and their expression levels in two mouse tissues and two cultured cell types, a list of five selected genes with lengths of ca. 100 kb and expression levels of ca. 1,000 TPM and a list of long lowly expressed genes used for comparison of cis-to-trans contact ratios with the studied genes.
Supplementary Table 3
Excel spreadsheet includes (1) the list and schematics of genomic BAC probes used for DNA and RNA FISH experiments; (2) primer pairs used for introduction of protospacer sequences into U6-gRNA-GFP-H2A; (3) primer sequences used to verify the expression of full length Ttn after activation with dCas9-VPR and the corresponding gel image; (4) primer pairs used to amplify Tg cDNA regions containing exons 2–12 and 33–47; (5) primers for generation of oligoprobes for Tg intron 41 (HD FISH); (6) primers for generation of oligoprobes for Tg 5′ exons and introns (SABER FISH); (7) primers for generation of probes for all Tg exons, intron 41 and the Sla gene; (8) primers for generation of probes for Ttn and Cald1 introns (SABER FISH).
Supplementary Table 4
Excel spreadsheet includes (1) the total number of acquired nuclei from five tissues and two cultured cell types, as well as the number of nuclei with a single expressed allele; (2) information on the number of reads in Hi-C experiments.
Supplementary Video 1
Confocal stacks through nuclei of mouse thyrocytes (counterstained with DAPI, red) after RNA FISH with a genomic probe for the Tg gene (green). Note the volatile shape of the Tg TLs and their great expansion into the nuclear interior.
Supplementary Video 2
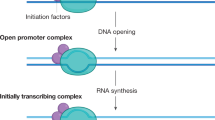
Cartoon showing how transcription initiation and termination of a highly expressed gene lead to formation and disappearance of a TL. The sequence of events: a gene body (orange thread) is coiled within a locus (blue threads) in a compact structure; upon transcription initiation, RNAPIIs (dark-grey oval structures) are loading at the gene promoter (red) and begin to elongate; during elongation, nRNPs appear and grow in size (depicted as grey amorphous structures); during a transcription pause, chromatin of the gene is coiled and forms a sliding knot (orange thread), dynamically formed beyond the last RNAPII of the first burst and disentangled by first RNAPII of the second burst; dense decoration with voluminous nRNP rigidifies the gene axis and forces its expansion, as well as the divergence of gene flanks (blue threads); RNAPIIs of the first transcription burst reach the 3′ gene end, release attached nRNPs and dissociate from the gene; the sliding chromatin knot reaches the 3′ gene end; RNAPIIs stop loading at the promoter at the end of the second burst; gene condensation starts 5′-terminally; the gene flanks begin to converge; the gene returns to its coiled compact state.
Source data
Source Data Fig. 3
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 7
Statistical source data.
Rights and permissions
About this article
Cite this article
Leidescher, S., Ribisel, J., Ullrich, S. et al. Spatial organization of transcribed eukaryotic genes. Nat Cell Biol 24, 327–339 (2022). https://doi.org/10.1038/s41556-022-00847-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-022-00847-6
This article is cited by
-
Co-transcriptional gene regulation in eukaryotes and prokaryotes
Nature Reviews Molecular Cell Biology (2024)
-
Transcriptional condensates: a blessing or a curse for gene regulation?
Communications Biology (2024)
-
Chromatin alternates between A and B compartments at kilobase scale for subgenic organization
Nature Communications (2023)
-
Transcripts in the loop
Nature Reviews Molecular Cell Biology (2022)
-
The sight of transcription
Nature Cell Biology (2022)