Abstract
Histone variants and the associated post-translational modifications that govern the stemness of haematopoietic stem cells (HSCs) and differentiation thereof into progenitors (HSPCs) have not been well defined. H3.3 is a replication-independent H3 histone variant in mammalian systems that is enriched at both H3K4me3- and H3K27me3-marked bivalent genes as well as H3K9me3-marked endogenous retroviral repeats. Here we show that H3.3, but not its chaperone Hira, prevents premature HSC exhaustion and differentiation into granulocyte-macrophage progenitors. H3.3-null HSPCs display reduced expression of stemness and lineage-specific genes with a predominant gain of H3K27me3 marks at their promoter regions. Concomitantly, loss of H3.3 leads to a reduction of H3K9me3 marks at endogenous retroviral repeats, opening up binding sites for the interferon regulatory factor family of transcription factors, allowing the survival of rare, persisting H3.3-null HSCs. We propose a model whereby H3.3 maintains adult HSC stemness by safeguarding the delicate interplay between H3K27me3 and H3K9me3 marks, enforcing chromatin adaptability.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The ChIP–seq data, including the fastq data and processed wig files, have been uploaded to the Gene Expression Omnibus website. The accession number is GSE148750. Please refer to Supplementary Table 10 for detailed information. Source data are provided with this paper.
Code availability
All the code will be available on request, including but not limited to the following: identification of the DEGs, identification of significantly increased or decreased peaks for H3K27me3 and H3K9me3 enrichment, calculation of H3K9me3 at the ERV sites, calculation of ERV mRNA expression and motif analysis.
Change history
20 January 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41556-022-00851-w
References
Yu, V. W. C. et al. Epigenetic memory underlies cell-autonomous heterogeneous behavior of hematopoietic stem cells. Cell 167, 1310–1322 (2016).
Yu, X. et al. Chromatin dynamics during the differentiation of long-term hematopoietic stem cells to multipotent progenitors. Blood Adv. 1, 887–898 (2017).
Lara-Astiaso, D. et al. Chromatin state dynamics during blood formation. Science 345, 943–949 (2014).
Corces, M. R. et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet. 48, 1193–1203 (2016).
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010).
Hoelper, D., Huang, H., Patel, D. J. & Lewis, P. W. Structural and mechanistic insights into ATRX-dependent and -independent functions of the histone chaperone DAXX. Nat. Commun. 8, 1193 (2017).
Lewis, P. W., Elsaesser, S. J., Noh, K.-M., Stadler, S. C. & Allis, C. D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl Acad. Sci. USA 107, 14075–14080 (2010).
Ricketts, M. D. et al. Ubinuclein-1 confers histone H3.3-specific-binding by the HIRA histone chaperone complex. Nat. Commun. 6, 7711 (2015).
Rai, T. S. et al. Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol. Cell. Biol. 31, 4107–4118 (2011).
Pietras, E. M., Warr, M. R. & Passegué, E. Cell cycle regulation in hematopoietic stem cells. J. Cell Biol. 195, 709–720 (2011).
Nakamura-Ishizu, A., Takizawa, H. & Suda, T. The analysis, roles and regulation of quiescence in hematopoietic stem cells. Development 141, 4656–4666 (2014).
Elsässer, S. J., Noh, K.-M., Diaz, N., Allis, C. D. & Banaszynski, L. A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 (2015).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Guo, P. et al. Endothelial jagged-2 sustains hematopoietic stem and progenitor reconstitution after myelosuppression. J. Clin. Invest. 127, 4242–4256 (2017).
Elsaesser, S. J. & Allis, C. D. HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harb. Sym. Quant. Biol. 75, 27–34 (2010).
Wong, L. H. et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 (2010).
Cui, K. et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4, 80–93 (2009).
Adli, M., Zhu, J. & Bernstein, B. E. Genome-wide chromatin maps derived from limited numbers of hematopoietic progenitors. Nat. Methods 7, 615–618 (2010).
Schepers, K., Hsiao, E. C., Garg, T., Scott, M. J. & Passegué, E. Activated Gs signaling in osteoblastic cells altersthe hematopoietic stem cell niche in mice. Blood 120, 3425–3435 (2012).
Pietras, E. M. et al. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell 17, 35–46 (2015).
Grinenko, T. et al. Hematopoietic stem cells can differentiate into restricted myeloid progenitors before cell division in mice. Nat. Commun. 9, 1898 (2018).
Wen, D. et al. Genome editing a mouse locus encoding a variant histone, H3.3B, to report on its expression in live animals. Genesis 52, 959–966 (2014).
Pradhan, S. K. et al. EP400 deposits H3.3 into promoters and enhancers during gene activation. Mol. Cell 61, 27–38 (2016).
Butler, J. M. et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell 6, 251–264 (2010).
Chen, P. et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Gene Dev. 27, 2109–2124 (2013).
Martire, S. et al. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat. Genet. 51, 941–946 (2019).
Bernstein, B. E. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Sachs, M. et al. Bivalent chromatin marks developmental regulatory genes in the mouse embryonic germline in vivo. Cell Rep. 3, 1777–1784 (2013).
Zentner, G. E., Tesar, P. J. & Scacheri, P. C. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res. 21, 1273–1283 (2011).
Matsumura, Y. et al. H3K4/H3K9me3 bivalent chromatin domains targeted by lineage-specific DNA methylation pauses adipocyte differentiation. Mol. Cell 60, 584–596 (2015).
Zhu, Y., van Essen, D. & Saccani, S. Cell-type-specific control of enhancer activity by H3K9 trimethylation. Mol. Cell 46, 408–423 (2012).
Young, G. R. et al. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491, 774–778 (2012).
Yoshinobu, K. et al. Selective up-regulation of intact, but not defective env RNAs of endogenous modified polytropic retrovirus by the Sgp3 locus of lupus-prone mice. J. Immunol. 182, 8094–8103 (2009).
Karimi, M. M. et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8, 676–687 (2011).
Sheng, W. et al. LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174, 549–563 (2018).
Chiappinelli, K. B. et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162, 974–986 (2015).
Izquierdo-Bouldstridge, A. et al. Histone H1 depletion triggers an interferon response in cancer cells via activation of heterochromatic repeats. Nucleic Acids Res. 45, 11622–11642 (2017).
Cuellar, T. L. et al. Silencing of retrotransposons by SETDB1 inhibits the interferon response in acute myeloid leukemiaSETDB1 regulates the antiviral response in AML. J. Cell Biol. 216, 3535–3549 (2017).
Collins, P. L., Kyle, Shinkai, Y. & Oltz, E. M. The histone methyltransferase SETDB1 represses endogenous and exogenous retroviruses in B lymphocytes. Proc. Natl Acad. Sci. USA 112, 8367–8372 (2015).
Stavrou, S. & Ross, S. R. APOBEC3 proteins in viral immunity. J. Immunol. 195, 4565–4570 (2015).
Elli, E. M., Baratè, C., Mendicino, F., Palandri, F. & Palumbo, G. A. Mechanisms underlying the anti-inflammatory and immunosuppressive activity of ruxolitinib. Front. Oncol. 9, 1186 (2019).
Li, W. et al. Type I interferon-regulated gene expression and signaling in murine mixed glial cells lacking signal transducers and activators of transcription 1 or 2 or interferon regulatory factor 9. J. Biol. Chem. 292, 5845–5859 (2017).
Platanias, L. C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5, 375–386 (2005).
Passegué, E. & Ernst, P. IFN-α wakes up sleeping hematopoietic stem cells. Nat. Med. 15, 612–613 (2009).
Baldridge, M. T., King, K. Y. & Goodell, M. A. Inflammatory signals regulate hematopoietic stem cells. Trends Immunol. 32, 57–65 (2011).
King, K. Y. & Goodell, M. A. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat. Rev. Immunol. 11, 685–692 (2011).
Mascarenhas, J. & Hoffman, R. Ruxolitinib: the first FDA approved therapy for the treatment of myelofibrosis. Clin. Cancer Res. 18, 3008–3014 (2012).
Ueda, Y., Kondo, M. & Kelsoe, G. Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J. Exp. Med. 201, 1771–1780 (2005).
He, Q. et al. The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell 17, 273–286 (2015).
Zhang, W. et al. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348, 1160–1163 (2015).
Challen, G. A., Boles, N. C., Chambers, S. M. & Goodell, M. A. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-β1. Cell Stem Cell 6, 265–278 (2010).
Ng, S. W. K. et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540, 433–437 (2016).
Shlush, L. I. et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547, 104–108 (2017).
Bagger, F. O., Kinalis, S. & Rapin, N. BloodSpot: a database of healthy and malignant haematopoiesis updated with purified and single cell mRNA sequencing profiles. Nucleic Acids Res. 47, D881–D885 (2018).
Shi, L., Wen, H. & Shi, X. The histone variant H3.3 in transcriptional regulation and human disease. J. Mol. Biol. 429, 1934–1945 (2017).
Buschbeck, M. & Hake, S. B. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 18, 299–314 (2017).
Henikoff, S. & Smith, M. M. Histone variants and epigenetics. Cold Spring Harb. Perspect. Biol. 7, a019364 (2015).
Szenker, E., Ray-Gallet, D. & Almouzni, G. The double face of the histone variant H3.3. Cell Res. 21, 421–434 (2011).
McKittrick, E., Gafken, P. R., Ahmad, K. & Henikoff, S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl Acad. Sci. USA 101, 1525–1530 (2004).
Farrell, M. J. et al. HIRA, a digeorge syndrome candidate gene, is required for cardiac outflow tract septation. Circ. Res. 84, 127–135 (1999).
Schwartz, B. E. & Ahmad, K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Gene Dev. 19, 804–814 (2005).
Wen, D. et al. Histone variant H3.3 is an essential maternal factor for oocyte reprogramming. Proc. Natl Acad. Sci. USA 111, 7325–7330 (2014).
Gerber J.P. et al. Aberrant chromatin landscape following loss of the H3.3 chaperone Daxx in haematopoietic precursors leads to Pu.1-mediated neutrophilia and inflammation. Nat. Cell Biol. https://doi.org/10.1038/s41556-021-00774-y (2021).
Garber, M., Grabherr, M. G., Guttman, M. & Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 8, 469–477 (2011).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Zang, C. et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25, 1952–1958 (2009).
Chu, H.-P. et al. TERRA RNA antagonizes ATRX and protects telomeres. Cell 170, 86–101 (2017).
Acknowledgements
We thank M. Ramalho-Santos at the Lunenfeld–Tanenbaum Research Institute, Mount Sinai Hospital for providing the Hirafl/fl mice. We thank L. Banaszynski for generating the plasmid containing the H3.3A floxed allele. We thank B. Kunar for consultation in the bioinformatics analyses. Y.L. is a New York Stem Cell Foundation Druckenmiller Fellow. We thank the Starr Foundation Stem Cell Derivation laboratory at Weill Cornell Medicine for the use of the FACS Aria II/III and BD LRSII systems. We thank the Mount Sinai Dean CoREs Flow Cytometry for their timely help and discussions with flow cytometry analysis and FACS. We thank the Mount Sinai CCMS for help with the complete blood count measurements of mouse peripheral blood. This work was funded by the New York State Stem Cell Science Program (NYSTEM; contract C32581GG to D.W. and S.R.) and grant no. R01 GM129380-01 from the National Institutes of Health (to D.W.). S.R. was funded by the Ansary Stem Cell Institute, National Institute of Health (grant nos R35 HL150809, RC2 DK114777 and U01AI138329), Empire State Stem Cell Board NYSTEM (grant no. C030160), Daedalus Fund for Innovation and Selma and Lawrence Ruben Science to Industry Bridge Fund from Weill Cornell Medicine, Starr Foundation Stem Cell Core Project and initiatives TRI-SCI 2019-029. R.S. was funded by NYSTEM contract C32596GG. B.S.D. was supported by the National Heart Lung and Blood Institute (grant no. R01HL130826) and NYSTEM (grant no. C34052GG).
Author information
Authors and Affiliations
Contributions
S.R., D.W. and P.G. conceived the project. S.R. and P.G. analysed the data and wrote the paper. D.W. generated the H3.3B−/− and H3.3A floxed mice. D.W., P.G. and Y.L. maintained the mice colonies and bred the mice. P.G., Y.L. and D.W. performed the mouse experiments. P.G. performed the in vitro cell-culture, transplantation and in vivo experiments. R.L., J.G.B.D. and T.I. offered critical evaluations regarding HSC transplantation. A.W.D. and A.R. provided the H3.3 rescue plasmid. P.G. and Y.L. performed the ChIP–seq experiments for the H3.3B–HA HSPCs. P.G. and X.L. performed ChIP–seq experiments using small numbers of LKS cells after tamoxifen deletion in vitro and in vivo. T.Z., F.T. and J.X. contributed to the bioinformatics analysis and offered insightful discussions. P.G. analysed and interpreted the ChIP–seq results. B.-S.D. provided funding for the project. B.-S.D., J.S. and S.Z.J. offered critical insights. P.G. analysed the RNA-seq results. S.Z.J. and S.R. helped interpret the results and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.R. is the non-paid co-founder of Angiocrine Bioscience (San Diego). The other authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks Gerald de Haan, Yuin-Han Loh and the other, anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 H3.3A and H3.3B are redundant during steady state hematopoiesis.
a. the Fragments Per Kilobase of transcript per Million mapped reads (FPKM) of H3.3A and H3.3B in hematopoietic cells. LT-HSC, long-term hematopoietic stem cells; MPP, multipotent progenitor cells; CMP, common myeloid progenitor cells; GMP, granulocyte macrophage progenitor cells; MEPs, megakaryocyte erythroid progenitor cells. Mono, Monocytes; Mac, macrophages; DCs, dendritic cells; CD11b+Gr1+ cells, granulocytes. The number of dots indicated the number of biological replicates. b. The design of the H3.3Afl/fl allele and the H3.3BEYFP/mcherry knockin knockout allele. c. qRT-PCR quantification of H3.3A mRNA expression within LKS cells after tamoxifen treatment. TKO is Id1mut/mut; DKO mice. d-e, genotyping of Rosa26creERT2, H3.3A floxed, WT, and deletion allele; H3.3B-EYFP or H3,3-mcherry allele (n>3 biological replicates were used). f-g. Percentages and numbers of c-Kit+Sca1+Lin− (LKS) or c-Kit+Sca1−Lin− LK cells within control (+/+, H3.3A+/−, or H3.3B+/−), H3.3AiKO/iKO (AKO), and H3.3B−/− (BKO) mice. h. Percentages of LKS and LK cells among LinN cells of control, AKO, and BKO mice. i. percentages of lineage cells within BM or spleen of control, AKO, and BKO mice. j. The total number of cells within BM, spleen, and thymus of H3.3B+/− (Bhet) or BKO mice. k-l, The total number of lineage cells with BM and spleen or Bhet or BKO mice. m-n, Recovery after myelosuppression with sublethal total body irradiation at 600 cGy. m, Survival probability after total body irradiation. n. white blood cells (WBC), red blood cells (RBC) and platelets (PLT) in the PB of control or BKO mice. o. Trimethylation at histone 3 lysine 27 (H3K27me3) profiling at WT or BKO LKS cells. Genome-wide, there were 3925 regions with reduced H3K27me3 enrichment. p. 210 of the H3K27me3 reduced regions in BKO LKS cells colocalize with H3.3 enrichment. q-r. We also profiled the enhancer mark, H3K27ac in BKO or WT cells, and identified the regions with increased or decreased H3K27ac enrichment. Motifs within those regions are also shown. For panels a, c, f-l, the number of dots indicate the number of animals. For m, n, n=5 mice for control group and n=9 for BKO group. Error bars indicate standard error of mean. p-value is calculated using unpaired, 2-tailed t-test. For h, p-value for the data points at day 27 was calculated using unpaired, 2-tailed t-test. Numerical source data are provided in Source Data.
Extended Data Fig. 2 Adult inducible deletion of H3.3A, but not its chaperone Hira resulted in myeloid bias.
a. FPKM of total H3.3, calculated as the sum of H3.3A and H3.3B, Hira, and Daxx in hematopoietic cells. b. Survival probability of DKO or BKO mice following tamoxifen treatment. c. Representative flow cytometric plots for the gating of LKS cells, LT-HSCs (CD150+CD48− LKS), MPP3 (CD150−CD48+ LKS), MPP2 (CD150+CD48+ LKS), and ST-HSCs (CD150−CD48− LKS) for BKO, DKO, Hirafl/fl or HiraKO mice. d-e. The percentages and the total number of LT-HSCs, MPP3, ST-HSCs, and LKS cells. f. The total number of progenitor cells at 2-3 weeks after H3.3 or Hira deletion. GMP, CD16/32+CD34+LK; MEPs, CD16/32−CD34−LK- cells; CMP, CD16/32−CD34+LK. g. At week 2, 3, 4, or 7 after tamoxifen injection, the upregulation of GMP marker CD16/32 on LKS and LT-HSCs in DKO mice. h. BM, spleen, and thymus of BKO, DKO, Hirafl/fl or HiraKO mice. Note the BM anemia phenotype and enlarged spleen for DKO mice. On the right, the spleen weight and thymocytes numbers are shown for HiraKO mice. i. CBC for BKO, DKO, Hirafl/fl or HiraKO mice. j-k. the percentages of granulocytes or B cells in the spleen or BM. l-n, the percentages of CD4+, CD8+, and CD4+CD8+ cells within BM, spleen, and thymus of the indicated mice. o. Representative flow cytometric plots of the erythroid cell clusters II-V. p-q. the percentages and numbers of Ter119+, erythroid cells II, III, IV, and V within the BM and spleen of BKO, DKO, Hirafl/fl, and HiraKO mice. For panels 2a, c, e-n, p-q, the number of dots indicates the number of animals Error bars indicate standard error of mean (SEM). P-values are calculated using unpaired, two-tailed, t-test. ‘ns’, non-significant. Numerical source data and unprocessed gels are provided in Source Data.
Extended Data Fig. 3 Histology of hematopoietic organs demonstrated myeloid hyperplasia in H3.3DKO mice.
a. Hematoxylin and eosin (HE) staining of the BM epiphysis and diaphysis of BKO, DKO, Hirafl/fl, and HiraKO mice. Note the increased bone mass and reduced hematopoietic cells in the epiphysis region of DKO mice, compared with BKO mice. b-d. HE staining of spleen (b), liver (c), and thymus (d) for BKO, DKO, Hirafl/fl, and HiraKO mice. Note the significantly smaller white pup in the spleen of DKO, but not that of HiraKO mice; the infiltration of hematopoietic cells in the liver of DKO mice, but not that of HiraKO mice. For a-d, n=4 independent animals for each genotype. e. Quantification of the percentages of the white pulp area in the spleens of BKO and DKO mice. The number of dots indicates the number of independent animals per experiment. The experiment in a-e were repeated 2-3 times. Error bar indicates SEM. P-value was calculated using two-tailed t-test. f. HE staining of lung, heart, and kidney from BKO and DKO mice. There were leukocyte infiltrations in the heart and kidney of DKO mice. g. Qiff-quik staining of BMMNCs and PB of BKO and DKO mice. Upper panel, BMMNCs, note the presence of red blood cells and lymphocytes in the BKO mice; however, DKO mice demonstrated myeloid hyperplasia, erythroid hypoplasia, and Peudo-Pelger huet cells (white arrows). Middle and lower panels, PB. In BKO PB, there were lymphocytes, RBCs, and neutrophils; in DKO mice, the PB contains predominantly myeloid cells (myeloid hyperplasia), including nucleated red cells, hypogranular neutrophils, ring-shaped neutrophils, myelocytes, myelocyte with micronuclei, and hyperlobated megakaryocytes (stag-horn shaped). For g, scale bar is 50 μm for each figure panel. For f-g, n=3 biological samples were used. Numerical source data are provided in Source Data.
Extended Data Fig. 4 H3.3 maintains hematopoietic homeostasis and HSC repopulation.
a. Percentages of CD45.2+ cells at the week 0 and week 4 after tamoxifen injection; note the absence of DKO-CRA cells in the PB of recipients CD45.1+ recipients before tamoxifen induced deletion of H3.3A gene, suggesting a disadvantage of BKO HSPCs to repopulate the wild type BM in a competitive setting. Experiments were repeated twice; each with n=4 animals for each group. b. Survival probability of mice with different transplantation schemes. c. Representative flow cytometric plots for CD16/32+ LKS cells in the recipients transplanted with DKO or AKO BMMNCs. d. Percentages of B220+ B cells and CD11b+Gr1+ granulocytes in the spleen of recipients transplanted with DKO or AKO BMMNCs. e. Percentages of B220+ B cells, Gr1+ myeloid cells, and CD3+ T cells in the PB of recipients transplanted with DKO or AKO BMMNCs. f-g. Images showing the skin lesions of the DKO BMMNCs-transplanted recipient animals, at week 7 after H3.3A deletion. The frequency, onset, and treatment for such skin lesion is listed in g. h-k. Hematoxylin and eosin (H and E) staining of BM, spleen, liver, and lung of recipients transplanted with DKO or AKO BMMNCs (n=3 for DKO, n=4 for AKO). For the BM, note the hypocellularity and sclerosis at the epiphysis and metaphysis regions. For the spleen, note the purple pro-fibrotic fibrous structures and disappearances of white pups in the DKO mice. There was no apparent hematopoietic cell infiltration into the liver for DKO BMMNC cells transplanted mice. For a, p-value is calculated using two-way ANOVA. For panels d, e, the number of dots indicates the number of independent animals. Error bars indicate standard error of mean. For d, e, p-value is calculated using unpaired, two-tailed t-test; the number of dots equals to the number of biological replicates in d, e. Numerical source data are provided in Source Data.
Extended Data Fig. 5 H3.3 maintains HSC repopulation in a competitive setting.
a. Representative flow cytometric plots for the CD45.2+ LinN cells and LT-HSC at week 16 following tamoxifen injection. b. The percentage and total number of CD45.2+ LinN cells within the BM of recipient mice. c. The percentages of CD45.2+ cells within the BM, spleen, and thymus of recipient mice. d-e. The total numbers of stem and progenitor cells, and lineage positive cells within the BM of recipient mice. f-g. Representative flow cytometric plots for the lineage cells (CD11b+, CD11b+Gr1+, Gr1+, or B220+ cells) within the BM or spleen of recipient mice. h. Representative flow cytometric plots for lineage cells (CD4+, CD8+, or CD4+CD8+ double positive cells) within thymus or spleen. i-j. The percentages of lineage-specific cells within BM or spleen of the recipients. k-l, The percentages of lineage cells within thymus or spleen of the recipients. For all the panels, n=6 animals for AKOBhet group; n=6 animals for DKO group. Error bars represent standard error of the mean. p-value is calculated using unpaired, 2-tailed t-test.
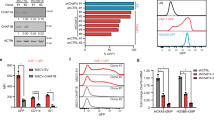
Extended Data Fig. 6 H3.3 prevents HSC myeloid differentiation.
a. Representative flow cytometric plots for the kinetics of LKS cell at different time points after tamoxifen treatment. Note the emergence of CD16/32+ LKS cells with the increase of culture time, and the elevated percentages of CD16/32+ cells in DKO LKS cells, compared with BKO LKS cells. b. percentages of the CD16/32+ among LK or LKS cells. c. Representative flow cytometric plots for the LT-HSCs during the co-culture. d. Lineage differentiation potential of BKO and DKO cells. There is reduced capacity for DKO to differentiate into B220+ B cells; overexpressing H3.3 did not rescue this defect. e. We set up in vitro culture assays for Hirafl/fl and Rosa26creERT2+ ;Hirafl/fl (HiraKO) HSCs. The growth curve of total hematopoietic cells are shown. f. the growth kinetics of total number of CD16/32+LKS cells. g. Representative flow cytometric plots for the LKS cells, LT-HSCs, GMPs, and CD16/32+ LKS cells in Hirafl/fl or HiraKO cultures. h-i, at day 4 after Hira deletion, the percentages and total numbers of LK, CMPs, GMPs, MEPs, and CD16/32+ LKS cells. j-k. the percentages and the total numbers of lineage cells, including B220+ B cells, CD11b+Gr1+ early myeloid cells, and CD11b+Gr1+ granulocytes. For panels 6b, d, e-f, h-k, the number of dots indicate the number of independent biological samples per experiment. Error bars indicate standard error of mean (SEM). For panels d, h, and i, P-values were calculated using unpaired, two-tailed, t-test; for panels b, e, f, j, and k, p-value was calculated using two-way ANOVA. Numerical source data are provided in Source Data.
Extended Data Fig. 7 H3.3 enrichment colocalizes with active histone marks H3K4me3 and repressive histone mark H3K27me3 or H3K9me3 within HSPCs.
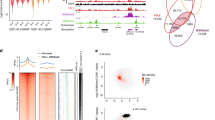
a. Schematic view of the H3.3B-HA-IRES-mcherry knockin mice22 used to map the H3.3 binding site within HSPCs. Lin− cells were co-cultured with E4-HUVECs for 1 week to expand LKS cells; ChIP-seq was performed using HA antibody. n=1 biological sample. b. Genome-wide distribution of 30,651 H3.3 peaks in LKS cells. Across the genome, there are 14,720 H3K4me3_H3.3 overlapped peaks (c), 2,012 H3.3 dependent H3K4me3/H3K27me3 bivalent peaks (d), and 3,484 H3K9me3_H3.3 overlapped peaks (f). e. The genome-wide localization of the H3.3 dependent H3K4me3/H3K27me3 regions. g. The genome-wide localization of the H3.3 dependent H3K9me3 marked regions. For panels c, d, e, f, ChIP-seq for histone modifications H3Km4e3, H3K9me3, H3K27me3 has been performed on n=3 biological samples. h. Genes at or near the H3.3-dependent H3K4me3/H3K27me3 bivalent regions. i. Genes at or near the H3.3 dependent H3K9me3 marked regions. j. There are a total of 9,031 genes with H3.3-dependent H3K4me3 enrichment, 2,533 genes with H3.3-dependent H3K9me3 enrichment, and 1,722 genes with H3.3-dependent H3K4me3/H3K27me3 enrichment. The expression levels of these three categories of genes are shown for both LT-HSCs and LKS cells. p-values were calculated using two-tailed t-test using one biological sample for each cell type. The experiment was repeated three times. The boundaries of box indicate the 25 and 75 percentiles, center indicate median number, whiskers indicate 1.5X interquartile range. k. Representative genome browser view of enrichment of H3.3, H3K4me3, H3K9me3, H3K27me3, H3K27ac and mRNA expression levels for active genes, bivalent genes and repetitive elements. Also shown here is the genome browser view of H3.3 enrichment using anti-H3.3 antibody, kindly shared by another group. P-value is labelled on the top of the box plot in panel j.
Extended Data Fig. 8 Dynamic changes of transcriptomic landscape within HSPCs following H3.3 deletion in vitro and in vivo.
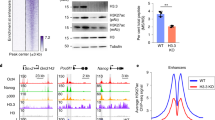
a-b, DAVID gene ontology biological pathway of differentially expressed genes (DEGs) at day 14-post H3.3 deletion in DKO LKS cells, compared with BKO LKS cells (in vivo). c-d, DAVID gene ontology biological pathway analysis of DEGs at day 56 post H3.3 deletion in DKO LKS cells, compared with BKO LKS cells (in vivo). e-f, the mean tag density of H3K4me3 and H3K9me3 at the promoter regions of the 385 downregulated genes associated with PC1. P-values were calculated using unpaired two-tailed t-test using one biological sample of each genotype. The experiment was performed twice independently for d14, once for d49, d56, and once for Hirafl/fl and HiraKO. g. Representative genome browser view of H3K4me3 and H3K27me3 intensity at intergenic region, at day 14 and day 56 post H3.3A deletion. h-k. For the commonly upregulated genes at d14 and d56 (171 genes), the enrichment of histone modifications H3K27me3, H3K27ac, H3K4me3, and H3K9me3 at the promoter regions is shown. For panels h-k, P-values were calculated using unpaired two-tailed t-test using one biological sample of each genotype. The experiment was performed twice independently for d14, once for d49, d56, and once for Hirafl/fl and HiraKO. l. Downregulation of key genes involved in erythrocyte differentiation within DKO LKS cells, compared with BKO LKS cells (n=2 biological samples). m, Venn diagram showing the overlap between the H3K27me3-reduced peaks within H3.3DKO cells and HiraKO LKS cells. n. The genome-wide distribution of DKO only, HiraKO and H3.3DKO shared, and HiraKO only H3K27me3-reduced peaks. o. The genes nearby the H3K27me3_reduced peaks are enriched in distinct biological pathways, as shown in the table. p. The H3K27me3 enrichment around the promoter regions of Id1 gene in Hirafl/fl or HiraKO LKS cells. For 8l, error bar indicates standard error of the mean. For the box plots in panels e-f, h-k, the boundaries of box indicate the 25 and 75 percentiles, center indicate median number, whiskers indicate 1.5X interquartile range. For e,-f, h-k, and o, p-values were calculated using two-tailed t-test. P-value is indicated on top of the box plot in panels e and f.
Extended Data Fig. 9 Dysregulated ERV expressions in H3.3DKO LKS cells triggered interferon responses, responsible for the myeloid bias.
a. H3K9me3 enrichment at representative ERV family. p-value was calculated using unpaired two-tailed t-test using one biological sample for each genotype. The experiment was repeated twice independently. b. The dynamic changes of ERV mRNA expression at in vitro or in vivo settings with DKO and BKO LKS cells (n=1 biological sample for each column). For the box plots in a, b, the boundaries of box indicate the 25 and 75 percentiles, center indicate median number, whiskers indicate 1.5X interquartile range. c. Genome browser plots showing the reduction of H3K9me3 and increased mRNA expression at a IAPEz-int ERV region. d. qRT-PCR quantifications of ERV expression at in vivo or in vitro LKS cells. env encodes for the envelope protein of ERV1. The number of dots indicate the number of biological samples]. e. At day 4 after in vitro deletion of H3.3A gene, the biological pathways associated with upregulated DEGs. f. We performed ChIP-seq for H3K27ac, an enhancer mark. The motifs within the H3K27ac increased regions include interferon pathway downstream transcription factor, Irf. g. The experimental scheme to test the effect of Jak2 inhibitor Ruxolitinib on the expansion and differentiation of HSPCs. h. Representative flow cytometric plots for the LKS, CD16/32+ LKS cells, pre-MegE, and pre-GM populations within the cocultures. i. The percentages of lineage positive cells within the total cultured cells. CD11b+Gr1+, immature neutrophils; CD11b+Gr1high, mature neutrophils. j. Representative flow cytometric plots for the lineage cells within the cocultures. k-m, at in vitro cultures, the mRNA expression of H3.3A, Hira, and Daxx within LKS cells. n-o, Daxx or Hira mRNA expression within LKS cells, GMPs, and LinP cells at in vivo scenarios. p-r, another batch of in vivo experiments, the mRNA expression of H3.3A, Hira, and Daxx within LKS cells was quantified. For panels I, k-m, o-s, the number of dots indicate the number of independent biological samples . Error bars indicate SEM. p-value is calculated using unpaired, two-tailed t-test unless otherwise indicated on the plot. P-value for panel a is labelled in the legend; p-value for panel b is not shown. Numerical source data are provided in Source Data.
Extended Data Fig. 10 Reduction of H3K9me3 opens up the accessibility of transcription factor binding sites.
a. Heatmap showing the increased ERV RNA expression within DKO LKS cells, compared with BKO LKS cells in chromosome 1. b. Heatmap showing the decreased ERV RNA expression within DKO cells, compared with BKO LKS cells. c. There is no H3K9me3 reduction mountain in HiraKO LKS cells. d. The H3K9me3 mountain in DKO cells persist into later stage d56 after H3.3A deletion. e. Karyotype plot showing the localization of H3K9me3 mountain in BKO LKS cells (blue bars), reduced H3K9me3 mountain in D4 DKO LKS cells (in vitro) (red triangle), and commonly reduced H3K9me3 mountains within three biological replicates. f. Karyotype plot showing the localization of H3K9me3 mountain in BKO LKS cells (blue bars), reduced H3K9me3 mountain in DKO LKS cells (red triangle), and commonly reduced H3K9me3 mountains within LKS and LK cells. g. The increased mRNA expression near decreased H3K9me3 peaks (distance from TSS < 10Kb) (n=2 biological samples). h. GMP marker Fcgr2b mRNA is upregulated in DKO HSPCs. Near Fcgr2b gene, there is increased H3K9me3 enrichment at ERV site, putative IRF binding site and Terra binding site67. i. Heatmap showing the expression of interferon target genes in BKO or DKO LKS cells, BKO or DKO GMPs. The gene with increased mRNA expression in DKO LKS cells compared with BKO LKS cells were labelled in bold. j, example of reduced mRNA expression near the H3K9me3_reduced_ERV regions. k-m. Kaplan Meyer survival curve for acute myeloid leukaemia patients striated according to the mRNA expression level of RELB (k), SETDB1 (k), and TRIM28 (l)54. For k and m, p-values were calculated using log-rank test. Error bars represent SEM. Numerical source data and unprocessed blots are provided in Source Data.
Supplementary information
Supplementary Table 1
Twelve supplementary tables in a single file consisting of 12 sheets. Supplementary Table 1. CBC for DKO, Bhet and BKO mice. Supplementary Table 2. CBCs for DKO and AhetBhet-transplanted mice. Supplementary Table 3. In vivo DEGs on d21. Supplementary Table 4. In vivo DEGs on d56. Supplementary Table 5. CBCs for DKO, BKO and TKO mice. Supplementary Table 6. H3K9me3 for DKO and BKO cells. Supplementary Table 7. In vivo DEGs on d14. Supplementary Table 8. FPKM for H3.3, H3.2 and H3.1 mRNA. Supplementary Table 9. FPKM for interferon pathway-related genes. Supplementary Table 10. High-throughput sequencing metadata template (version 2.1). Supplementary Table 11. Primers used in the study. Supplementary Table 12. Antibodies used in this study and their dilutions.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Figure. 1
Unprocessed DNA gel electrophoresis.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Figure. 9
Unprocessed western blots.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Guo, P., Liu, Y., Geng, F. et al. Histone variant H3.3 maintains adult haematopoietic stem cell homeostasis by enforcing chromatin adaptability. Nat Cell Biol 24, 99–111 (2022). https://doi.org/10.1038/s41556-021-00795-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-021-00795-7
This article is cited by
-
KAT2A-mediated succinylation modification of notch1 promotes the proliferation and differentiation of dental pulp stem cells by activating notch pathway
BMC Oral Health (2024)
-
HIRA vs. DAXX: the two axes shaping the histone H3.3 landscape
Experimental & Molecular Medicine (2024)
-
The Role of H3K27me3-Mediated Th17 Differentiation in Ankylosing Spondylitis
Inflammation (2024)
-
The histone chaperone function of Daxx is dispensable for embryonic development
Cell Death & Disease (2023)
-
Transcription factor Nkx2-3 maintains the self-renewal of hematopoietic stem cells by regulating mitophagy
Leukemia (2023)