Abstract
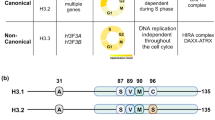
The histone variant H3.3 is enriched at enhancers and active genes, as well as repeat regions such as telomeres and retroelements, in mouse embryonic stem cells (mESCs)1,2,3. Although recent studies demonstrate a role for H3.3 and its chaperones in establishing heterochromatin at repeat regions4,5,6,7,8, the function of H3.3 in transcription regulation has been less clear9,10,11,12,13,14,15,16. Here, we find that H3.3-specific phosphorylation17,18,19 stimulates activity of the acetyltransferase p300 in trans, suggesting that H3.3 acts as a nucleosomal cofactor for p300. Depletion of H3.3 from mESCs reduces acetylation on histone H3 at lysine 27 (H3K27ac) at enhancers. Compared with wild-type cells, those lacking H3.3 demonstrate reduced capacity to acetylate enhancers that are activated upon differentiation, along with reduced ability to reprogram cell fate. Our study demonstrates that a single amino acid in a histone variant can integrate signaling information and impact genome regulation globally, which may help to better understand how mutations in these proteins contribute to human cancers20,21.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
Code to generate figures is available at https://github.com/utsw-medical-center-banaszynski-lab/Martire-et-al-2019-Nature-Genetics.git
References
Shi, L., Wen, H. & Shi, X. The histone variant H3.3 in transcriptional regulation and human disease. J. Mol. Biol. 429, 1934–1945 (2017).
Goldberg, A. D. et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140, 678–691 (2010).
Wong, L. H. et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20, 351–360 (2010).
Banaszynski, L. A. et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 155, 107–120 (2013).
Elsässer, S. J., Noh, K.-M., Diaz, N., Allis, C. D. & Banaszynski, L. A. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522, 240–244 (2015).
Sadic, D. et al. Atrx promotes heterochromatin formation at retrotransposons. EMBO Rep 16, 836–850 (2015).
He, Q. et al. The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell 17, 273–286 (2015).
Voon, H. P. J. et al. ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep. 11, 405–418 (2015).
Jang, C.-W., Shibata, Y., Starmer, J., Yee, D. & Magnuson, T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 29, 1377–1392 (2015).
Hödl, M. & Basler, K. Transcription in the absence of histone H3.3. Curr. Biol. 19, 1221–1226 (2009).
Sakai, A., Schwartz, B. E., Goldstein, S. & Ahmad, K. Transcriptional and developmental functions of the H3.3 histone variant in Drosophila. Curr. Biol. 19, 1816–1820 (2009).
Kim, H., Heo, K., Choi, J., Kim, K. & An, W. Histone variant H3.3 stimulates HSP70 transcription through cooperation with HP1γ. Nucleic Acids Res. 39, 8329–8341 (2011).
Tamura, T. et al. Inducible deposition of the histone variant H3.3 in interferon-stimulated genes. J. Biol. Chem. 284, 12217–12225 (2009).
Zhang, H. et al. RPA interacts with HIRA and regulates H3.3 deposition at gene regulatory elements in mammalian cells. Mol. Cell 65, 272–284 (2017).
Nashun, B. et al. Continuous histone replacement by hira is essential for normal transcriptional regulation and de novo DNA methylation during mouse oogenesis. Mol. Cell 60, 611–625 (2015).
Kong, Q. et al. Histone variant H3.3-mediated chromatin remodeling is essential for paternal genome activation in mouse preimplantation embryos. J. Biol. Chem. 293, 3829–3838 (2018).
Wong, L. H. et al. Histone H3.3 incorporation provides a unique and functionally essential telomeric chromatin in embryonic stem cells. Genome Res. 19, 404–414 (2009).
Hake, S. B. et al. Serine 31 phosphorylation of histone variant H3.3 is specific to regions bordering centromeres in metaphase chromosomes. Proc. Natl Acad. Sci. USA 102, 6344–6349 (2005).
Chang, F. T. M. et al. CHK1-driven histone H3.3 serine 31 phosphorylation is important for chromatin maintenance and cell survival in human ALT cancer cells. Nucleic Acids Res. 43, 2603–2614 (2015).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Behjati, S. et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat. Genet. 45, 1479–1482 (2013).
Pasini, D. et al. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res. 38, 4958–4969 (2010).
Chen, X. et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 (2008).
Pradeepa, M. M. et al. Histone H3 globular domain acetylation identifies a new class of enhancers. Nat. Genet. 48, 681–686 (2016).
Dorighi, K. M. et al. Mll3 and Mll4 facilitate enhancer RNA synthesis and transcription from promoters independently of H3K4 monomethylation. Mol. Cell 66, 568–576.e4 (2017).
Rickels, R. et al. Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 49, 1647–1653 (2017).
Lai, B. et al. MLL3/MLL4 are required for CBP/p300 binding on enhancers and super-enhancer formation in brown adipogenesis. Nucleic Acids Res. 45, 6388–6403 (2017).
Wang, C. et al. Enhancer priming by H3K4 methyltransferase MLL4 controls cell fate transition. Proc. Natl Acad. Sci. USA 113, 11871–11876 (2016).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Josefowicz, S. Z. et al. Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol. Cell 64, 347–361 (2016).
Cheung, P. et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5, 905–915 (2000).
Li, M., Dong, Q. & Zhu, B. Aurora kinase B phosphorylates histone H3.3 at serine 31 during mitosis in mammalian cells. J. Mol. Biol. 429, 2042–2045 (2017).
Delvecchio, M., Gaucher, J., Aguilar-Gurrieri, C., Ortega, E. & Panne, D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat. Struct. Mol. Biol. 20, 1040–1046 (2013).
Ortega, E. et al. Transcription factor dimerization activates the p300 acetyltransferase. Nature 562, 538–544 (2018).
Thorne, J. L., Ouboussad, L. & Lefevre, P. F. Heterochromatin protein 1 gamma and IκB kinase alpha interdependence during tumour necrosis factor gene transcription elongation in activated macrophages. Nucleic Acids Res. 40, 7676–7689 (2012).
Shimada, M. et al. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell 132, 221–232 (2008).
Guo, R. et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell 56, 298–310 (2014).
Wen, H. et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature 508, 263–268 (2014).
Roberts, C. et al. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell. Biol. 22, 2318–2328 (2002).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Jiang, L. et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 21, 1543–1551 (2011).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Acknowledgements
We thank members of the Banaszynski and Hon laboratories for helpful discussions; E. Duncan, H. Yu, L. Kraus, H. Zhu, and S. Morrison for critical comments on this manuscript; A. Shoshnev for the illustrations used as templates in this manuscript; UTSW BioHPC for computational infrastructure; UTSW McDermott Center and the Children’s Research Institute for providing next-generation sequencing services; UTSW Flow Cytometry Core for single cell isolation; UTSW Proteomics Core for mass spectrometry analysis. L.A.B. is a Virginia Murchison Linthicum Scholar in Medical Research (UTSW Endowed Scholars Program) and a Peterson Investigator of the Neuroendocrine Research Foundation (NETRF). This work was supported in part by CPRIT RR140042, The Welch Foundation I-1892, DoD KCRP KC170230, and NIH R35 GM124958 (L.A.B.), the American-Italian Cancer Foundation (S.M.), the Taiwan Postdoctoral Research Abroad Fellowship (Y.-C.T), and the Green Center for Reproductive Biology Sciences.
Author information
Authors and Affiliations
Contributions
S.M. and L.A.B. conceived and designed the study; S.M. performed experiments with help from A.T., J.N., Y.-C.T., and M.T.; A.A.G. performed computational analyses; A.W. contributed to the differentiation study; L.A.B. supervised the project; S.M. and L.A.B. wrote the manuscript with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Histone H3.3 facilitates histone acetylation at enhancers and promoters.
Related to Fig. 1. a, Average profile showing enrichment of H3 and H3.3 at promoter-distal enhancers (± 3 kb from TSS) in mESC, related to Fig. 1a. b, Immunoblot of whole cell lysates from WT and H3.3 KO2 (independent clone) mESCs. Blot is representative of three independent experiments. See also Supplementary Fig. 10 for all the blots in this figure. c, Immunoblot with the indicated antibodies of whole cell lysates from WT and H3.3 KO mESCs. Blot is representative of three independent experiments. d, Dot blot array showing H3K27ac antibodies reactivity for either H3.1K27ac or H3.3K27ac peptides. Blot is representative of two independent experiments. e, Quantification of histone acetylation and methylation levels in WT and H3.3 KO mESCs by mass spectrometry (Mod SpecTM, Active Motif). Relative abundance across samples is reported. Data represented as mean ± s.d (n = 3). f, Bar graph showing the number of H3K27ac peaks that show changes in H3K27ac levels between WT and H3.3 KO mESC at enhancers (± 3 kb from TSS). g, Boxplots showing H3K27ac enrichment levels for distal enhancers that show reduced (n = 13,135) and increased (n = 2,454) H3K27ac enrichment in H3.3 KO mESCs compared to WT. The bottom and top of the boxes correspond to the 25th and 75th percentiles, and the internal band is the 50th percentile (median). The plot whiskers correspond to 1.5× interquartile range and outliers are excluded. h, Average profile of H3K27ac enrichment at enhancers in WT and H3.3 KO mESCs using a monoclonal antibody. i,j, ChIP-qPCR validation from independent experiments of H3K27ac enrichment at 16 enhancers in WT and 2 clonal H3.3 KO mESCs. k, Ratio (log2) of H3K27ac enrichment in WT versus H3.3 KO mESCs at promoters and enhancers showing reduced acetylation in the absence of H3.3. x axis values > 0 indicate reduced H3K27ac enrichment in the absence of H3.3. l,m,n, Average profile of H3K18ac (l) H3K64ac (m) and H3K122ac (n) enrichment at enhancers in WT and H3.3 KO mESCs. o,p, Average profile (o) and ChIP-qPCR validation (p) of H3K4me1 enrichment at enhancers in WT and H3.3 KO mESCs.
Supplementary Figure 2 H3K27ac at enhancers is facilitated by HIRA-dependent H3.3 deposition.
Related to Fig. 1. a, Immunoblot with the indicated antibodies of whole cell lysates from WT and HIRA, ATRX and DAXX KO mESCs. Blot is representative of three independent experiments. See also Supplementary Fig. 11 for all the blots in this figure. b, Boxplot showing H3.3 enrichment at enhancers in WT and HIRA, ATRX and DAXX KO mESCs (n = 17,589). WT & HIRA KO P < 2.2 × 10−16, WT & ATRX KO P = 1.664 × 10−14, WT & DAXX KO P = 0.0007246 by Wilcoxon rank sum two-side test. The bottom and top of the boxes correspond to the 25th and 75th percentiles, and the internal band is the 50th percentile (median). The plot whiskers correspond to 1.5× interquartile range and outliers are excluded. c, Immunoblot with the indicated antibodies from WT and HIRA KO mESCs whole cell lysates loaded with increased protein concentration (1x, 2x). Blot is representative blot of two independent experiments. d, Average profile of H3K27ac enrichment at enhancers in WT and HIRA KO mESCs. e, Venn diagram showing overlap between H3.3 KO and HIRA KO reduced H3K27ac peaks. f, Average profile of H3K27ac enrichment at enhancers in WT, ATRX KO, and DAXX KO mESCs. g, Schematic of CRISPR/Cas9 strategy for generation of HIRA KO mESCs and immunoblot showing complete loss of HIRA. Blot is representative of four independent experiments. h, Average profile of H3K27ac enrichment in WT and HIRA KO2 mESCs at distal enhancers. i, Venn diagram showing overlap between HIRA KO and HIRA KO2 reduced H3K27ac peaks. j, ChIP-qPCR validation from independent experiments of H3K27ac enrichment at enhancers in all four chaperone KO mESCs line compared to the corresponding WT mESCs.
Supplementary Figure 3 H3.3 loss does not alter chromatin accessibility or p300 recruitment.
Related to Fig. 2. a, Quantification of Acetyl-CoA levels from 5×106 cells (Sigma MAK039) in WT and H3.3 KO mESC. Data represented as mean ± s.d. (n = 3), two-tailed t-test, n.s. = 0.9. b, HDAC activity (BioVision K331-100) in WT and H3.3 KO mESC nuclear extracts in the presence or absence of the HDAC inhibitor Sodium Butyrate (NaBut). Data represented as mean ± s.d. (n = 5), two-tailed t-test, n.s. = 0.45. c, Immunoblot of cytoplasmic (C) and nuclear extracts (NE) from WT and H3.3 KO mESCs. Blot is representative of three independent experiments. See also Supplementary Fig. 11b. d, Immunoblot of whole cell lysates from WT mESCs transfected with p300 and CBP shRNA. Blot is representative of four independent experiments. See also Supplementary Fig. 11b. e, Heatmaps and average profiles of H3K27ac (left), ATAC-seq (middle), and p300 (right) enrichment in WT and H3.3 KO mESCs at enhancers. 3kb around the center of enhancers are displayed for each analysis. Each row represents a single enhancer (n =17,589). f, Heatmap and average profile of ATAC-seq in WT and HIRA KO mESCs at enhancers. 3kb around the center of enhancers are displayed for each analysis. Each row represents a single enhancer (n = 17,589).
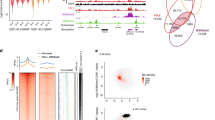
Supplementary Figure 4 H3.3 phosphorylation promotes acetylation at enhancers.
Related to Fig. 2. a, Boxplots showing H3-Flag-HA (left) and H3K27ac (right) enrichment (ChIP-qPCR) at 10 enhancer regions in H3.3 KO mESCs expressing exogenous histones (H3.2 and H3.3) compared to WT mESCs. (n = 10). The bottom and top of the boxes correspond to the 25th and 75th percentiles, and the internal band is the 50th percentile (median). The plot whiskers correspond to 1.5× interquartile range and outliers are excluded. b, Immunoblot of whole cell lysates from WT and H3.3 KO mESCs exogenously expressing histones and indicated histone mutants. Blot is representative of two independent experiments. See also Supplementary Fig. 12a for all the blots in this figure. c, Venn diagrams showing drastically reduced H3.3S31ph peaks in WT mESC treated with Chek1inhibitor (Chk1i). p = 6.419 x 10-14 by hypergeometric test. Ctrl peaks (n = 11,259), Cheki peaks (n = 533). d, Violin plot showing H3.3S31ph in WT and mESC treated with Chek1 inhibitor (Chk1i). ***P < 2.2 × 10−16 by Wilcoxon rank sum two-side test (n = 11,259). Violin plots displayed as boxplot in panel a. e, Genome-wide distribution of H3.3S31ph binding sites (percent of total number of sites. H3.3S31ph ChIP-seq identified elements that fell within 300 kb from coding regions were analyzed based on their distance from nearest genes using CEAS. The pie chart represents percent distribution. f, ChIP-qPCR of H3.3S31ph enrichment at enhancers in WT and HIRA2 KO mESCs. Data are expressed as percentage of input and represent mean ± s.d. (n = 8), two-tailed t-test **P = 0.0016. g, Immunoblot of whole cell lysates from WT mESCs treated with indicated concentration of Chk1 kinase inhibitor (SB218078, ‘SB’) or Aurora B kinase inhibitor (ZM447439, ‘ZM’) for 4 hrs. Blot is representative of three independent experiments. h, Immunoblot of whole cell lysates from wild-type mESCs treated with Chk1 inhibitor for the indicated times. Blot is representative of three independent experiments.
Supplementary Figure 5 H3.3-specific phosphorylation stimulates p300 histone acetyltransferase activity.
Related to Fig. 3. a-b, Immunoblot (above) and densitometry (below) of p300 histone acetyltransferase assay using increasing concentrations of recombinant H3.3 (a) or H3.1 (b) nucleosomes that were preincubated with Chk1 with or without ATP. Blot is representative of three independent experiments. See also Supplementary Fig. 12b.
Supplementary Figure 6 mESCs depleted of H3.3 maintain self-renewal properties.
Related to Fig. 4. a,c,e, MA plot of gene expression in WT and H3.3 KO (a), HIRA KO1 (c), HIRA KO2 (e) mESCs. The x-axis indicates gene counts and the y-axis represents the log2 fold change in expression for KO versus WT mESCs. Genes in red and blue were differentially expressed (P < 0.05) with a fold change > 2. b, Venn diagrams showing the relationship between H3K27ac ChIP-seq and RNA-seq for regions that showed upregulation (top) or downregulation (bottom) in the absence of H3.3 in both data sets. Up regulated genes P = 6.419 × 10−14 and down regulated genes P = 3.581 × 10−12 by hypergeometric test. H3K27ac Up-regulated (n = 1,051), RNA-Seq Up-regulated (n = 824), H3K27ac Down-regulated (n = 2,796), RNA-Seq Down-regulated (n = 724). d,f, Venn diagrams showing the relationship between H3K27ac ChIP-seq and RNA-seq for regions that showed downregulation in the absence of H3.3 in HIRA KO cells. Down regulated genes P = 3.581 × 10-12 by hypergeometric test. (d) H3K27ac Down-regulated (n = 2,022), RNA-Seq Down-regulated (n = 704). (f) H3K27ac Down-regulated (n = 1736), RNA-Seq Down-regulated (n = 859). g, Alkaline phosphatase staining of WT and H3.3 KO mESCs in 2i/SL media. Scale bar = 200 μm. Image is representative of three independent experiments. h, Quantification of colony formation assay of WT and H3.3 KO mESCs. i, Growth curve of WT and H3.3 KO mESCs assessed using MTT assay at indicated times. Data represent mean ± s.d (n = 4).
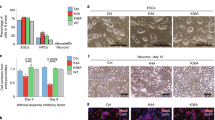
Supplementary Figure 7 H3.3 Is required for mESC differentiation.
Related to Fig. 4. a, Average profiles of H3.3, H3K4me1, H3K27ac and p300 enrichment in WT mESC at mESC and EBs enhancers. 3kb around the center of enhancers are displayed for each analysis. b, Brightfield images of embryoid body (EB) differentiation in WT and H3.3 KO cells at 4, 8 and 12 days. Scale bar = 1 mm. Image is representative of at least three independent experiments. c, Comparison of average areas of WT and H3.3 KO embryoid bodies at the indicated days of differentiation. Data represent the mean ± s.d., two-tailed t-test ***P = 7.52−53, **P = 0.0068, ***P = 0.00016. d, Heatmap representation of transcript levels (RT-qPCR) of markers of pluripotency (ESC) (Oct4, Nanog), mesoderm (Brachyury, Fgf5), endoderm (Gata4, Gata6) and ectoderm (Cdx2, Otx6) during EBs differentiation. e, Comparison of transcription changes at day 4 of EB differentiation for WT and H3.3 KO cells. Gene list represent genes whose expression decreases or increases in expression upon differentiation compared to WT ESCs. The x- and y-axis represent the fold change (log2) of RNA expression between D4 and D0 in WT and H3.3 KO, respectively. f, Immunoblot of H3.3 and Oct4 in WT and H3.3 KO embryoid bodies at the indicated days of differentiation. Blot is representative of two independent experiments. See also Supplementary Fig. 12c. g, Model figure representing differential requirement for H3.3 during ongoing transcription (left) and the initiation of transcription (right).
Supplementary Figure 8 H3.3 phosphorylation promotes enhancer acetylation during differentiation.
Related to Fig. 4. a, Average profiles of H3K27ac enrichment at EB enhancers in H3.3KO EBs expressing the indicated exogenous protein. 3kb around the center of enhancers are displayed for each analysis. b, Brightfield images of EB differentiation in WT and H3.3 KO cells and in H3.3 KO cells expressing exogenous histone mutants at day 4. Scale bar = 1 mm. Image is representative of three independent experiments. c, Average profiles of H3K27ac enrichment at ESC enhancers in H3.3KO EBs expressing the indicated exogenous protein. 3kb around the center of enhancers are displayed for each analysis. d, Schematic of H3K27ac levels in WT and in H3.3 KO cells expressing exogenous histone mutants at day 4 of differentiation at mESC (pluripotency-specific) and EB (differentiation-specific) enhancers.
Supplementary Figure 10 Raw immunoblots.
a, Western blots related to Supplementary Fig. 1b–d.
Supplementary information
Supplementary Information
Supplementary Figures 1–12 and Supplementary Note
Rights and permissions
About this article
Cite this article
Martire, S., Gogate, A.A., Whitmill, A. et al. Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nat Genet 51, 941–946 (2019). https://doi.org/10.1038/s41588-019-0428-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-019-0428-5
This article is cited by
-
HIRA vs. DAXX: the two axes shaping the histone H3.3 landscape
Experimental & Molecular Medicine (2024)
-
H3.3 contributes to chromatin accessibility and transcription factor binding at promoter-proximal regulatory elements in embryonic stem cells
Genome Biology (2023)
-
Knockout tales: the versatile roles of histone H3.3 in development and disease
Epigenetics & Chromatin (2023)
-
Histone exchange sensors reveal variant specific dynamics in mouse embryonic stem cells
Nature Communications (2023)
-
Enduring questions in regenerative biology and the search for answers
Communications Biology (2023)