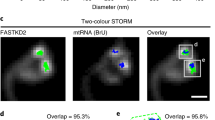
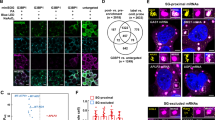
Abstract
Ribonucleoprotein (RNP) granules are non-membrane-bound organelles that have critical roles in the stress response1,2, maternal messenger RNA storage3, synaptic plasticity4, tumour progression5,6 and neurodegeneration7,8,9. However, the dynamics of their mRNA components within and near the granule surface remain poorly characterized, particularly in the context and timing of mRNAs exiting translation. Herein, we used multicolour single-molecule tracking to quantify the precise timing and kinetics of single mRNAs as they exit translation and enter RNP granules during stress. We observed single mRNAs interacting with stress granules and P-bodies, with mRNAs moving bidirectionally between them. Although translating mRNAs only interact with RNP granules dynamically, non-translating mRNAs can form stable, and sometimes rigid, associations with RNP granules with stability increasing with both mRNA length and granule size. Live and fixed cell imaging demonstrated that mRNAs can extend beyond the protein surface of a stress granule, which may facilitate interactions between RNP granules. Thus, the recruitment of mRNPs to RNP granules involves dynamic, stable and extended interactions affected by translation status, mRNA length and granule size that collectively regulate RNP granule dynamics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Code availability
Custom Mathematica (version 11.2.0.0) code was deposited on GitHub and can be accessed at https://raw.githubusercontent.com/TatsuyaMorisaki/Translation-Stress/master/Translation-Stress.nb.
Data availability
Source data for Figs. 1b−f, 2c–e, 3a–d, 4a,c–e and Supplementary Figs. 1,2,4 are provided in Supplementary Table 2. The raw images were deposited to figshare and can be accessed at: https://figshare.com/articles/Dataset_associated_with_Multicolor_single-molecule_tracking_of_mRNA_interactions_with_RNP_granules_-1_2/7427816 and https://figshare.com/articles/Dataset_associated_with_Multicolor_single-molecule_tracking_of_mRNA_interactions_with_RNP_granules_-2_2/7427918. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
References
Buchan, J. R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009).
Kedersha, N., Ivanov, P. & Anderson, P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem. Sci. 38, 494–506 (2013).
Schisa, J. A. Effects of stress and aging on ribonucleoprotein assembly and function in the germ line. Wiley Interdiscip. Rev. RNA 5, 231–246 (2014).
Sudhakaran, I. P. et al. FMRP and ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl Acad. Sci. USA 111, E99–E108 (2014).
El-Naggar, A. M. & Sorensen, P. H. Translational control of aberrant stress responses as a hallmark of cancer. J. Pathol. 244, 650–666 (2018).
Grabocka, E. & Bar-Sagi, D. Mutant KRAS enhances tumor cell fitness by upregulating stress granules. Cell 167, 1803–1813 (2016).
Zhang, K. et al. Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971 (2018).
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006).
Li, Y. R., King, O. D., Shorter, J. & Gitler, A. D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013).
Morisaki, T. et al. Real-time quantification of single RNA translation dynamics in living cells. Science 352, 1425–1429 (2016).
Grimm, J. B. et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–250 (2015).
Kedersha, N., Tisdale, S., Hickman, T. & Anderson, P. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448, 521–552 (2008).
Khong, A. et al. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808–820 (2017).
Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R. & Parker, R. Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 (2016).
Kedersha, N. et al. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151, 1257–1268 (2000).
Ohshima, D., Arimoto-Matsuzaki, K., Tomida, T., Takekawa, M. & Ichikawa, K. Spatio-temporal dynamics and mechanisms of stress granule assembly. PLoS Comput. Biol. 11, e1004326 (2015).
Namkoong, S., Ho, A., Woo, Y. M., Kwakm, H. & Lee, J. H. Systematic characterization of stress-induced RNA granulation. Mol. Cell 70, 175–187 (2018).
Wilbertz, J. H. et al. Single-molecule imaging of mRNA localization and regulation during the integrated stress response. Mol. Cell https://doi.org/10.1016/j.molcel.2018.12.006 (2019).
Kedersha, N. et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884 (2005).
Simpson, C. E., Lui, J., Kershaw, C. J., Sims, P. F. G. & Ashe, M. P. mRNA localization to P-bodies in yeast is biphasic with many mRNAs captured in a late Bfr1p-dependent wave. J. Cell Sci. 127, 1254–1262 (2014).
Wei, M.-T. et al. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 9, 1118–1125 (2017).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Molliex, A. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015).
Sfakianos, A. P., Whitmarsh, A. J. & Ashe, M. P. Ribonucleoprotein bodies are phased. Biochem. Soc. Trans. 44, 1411–1416 (2016).
Niewidok, B. et al. Single-molecule imaging reveals dynamic biphasic partition of RNA-binding proteins in stress granules. J. Cell Biol. 217, 1303–1318 (2018).
Banani, S. F. et al. Compositional control of phase-separated cellular bodies. Cell 166, 651–663 (2016).
Tokunaga, M., Imamoto, N. & Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008).
Edelstein, A. D. et al. Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10 (2014).
Edelstein, A., Amodaj, N., Hoover, K., Vale, R. & Stuurman, N. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. 29, 14.20.1–14.20.17 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Stellaris® RNA FISH: Protocol for Sequential IF + FISH in Adherent Cells (Biosearch Technologies, 2015); https://biosearchassets.blob.core.windows.net/assets/bti_custom_stellaris_immunofluorescence_seq_protocol.pdf.
Acknowledgements
We thank N. Kedersha for providing GFP–G3BP1/mRFP–DCP1a U-2 OS cells, L. Lavis for providing JF dyes, and E. Braselmann and T. Nahreini for isolating GFP–G3BP1 U-2 OS cells at the BioFrontiers Institute Flow Cytometry Core facility. We thank B. Dodd and lab members for assistance and helpful suggestions. S.L.M. was funded by the Anna and John J. Sie Foundation; A.K. by a Banting postdoctoral fellowship; R.P. by the HHMI; T.J.S. by the NIH (grant no. R35GM119728) and the Boettcher Foundation’s Webb-Waring Biomedical Research Program.
Author information
Authors and Affiliations
Contributions
S.L.M. and R.P. conceptualized the study; S.L.M., T.M., T.J.S., A.K. and K.L. developed and designed methodology; T.M. and T.J.S. developed and implemented software; S.L.M. validated findings; T.M., T.J.S., S.L.M. and A.K. performed formal analyses; S.L.M., A.K., K.L., T.M. and T.J.S. performed experiments and/or collected data; R.P., T.J.S., T.M., A.K., K.L. and S.L.M. provided resources; T.M., T.J.S. and S.L.M. provided data curation; S.L.M. and R.P. wrote the original draft; R.P., S.L.M., T.J.S., T.M. and A.K. reviewed and edited the drafts; T.M., T.J.S., S.L.M. and A.K. visualized data; R.P., T.J.S., T.M. and S.L.M. supervised the project; S.L.M. managed and coordinated the project; T.J.S., R.P. and S.L.M. acquired funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 Translating mRNAs can (rarely) interact with stress granules for several minutes.
A representative long-term interaction between a translating KDM5B mRNA and a SG (>2 min) from 2 mRNAs tracked from 2 cells from 2 independent experiments. The mRNA translation signal persists throughout the interaction with the SG and after exiting the SG. This mRNA is shown as “mRNA2” in Supplementary Movie 3
Supplementary Figure 2 Translation resumes following complete disassembly of stress granules in a single cell during recovery from stress.
Single-cell translation resumption during the recovery from arsenite stress (representative cell shown from 4 cells collected from 2 independent experiments). Translation resumes just a few minutes after SG disassembly. Number of translation foci (green) and SGs (blue) during stress (0–60 min) and following arsenite washout (70–130 min)
Supplementary Figure 3 The distance and angle between mRNAs within a single stress granule can remain consistent over time.
Quantification of the relative locations of KDM5B mRNAs shown in Fig. 4b and c, and Supplementary Movies 11 and 12. The distances (a) and the angles (b) between two KDM5B mRNAs interacting with the same SG are shown. The pair of mRNAs used for distance and angle calculations are shown with arrows (right). The average distances and angles are shown in the left top insets (± SD) from n = 300 time points, from 2 cells collected from 2 independent experiments. (c) Distance of one mRNA (red dot) from the surface of the stress granule (blue). This mRNA exits and re-enters the SG
Supplementary Figure 4 mRNAs can be rigidly adhered within a stress granule and may be tethered to the granule surface.
Representative time series showing three KDM5B mRNAs (depicted as red, blue or green arrows and dots) positioned within SGs over time (from 300 times points, representing one of 2 SGs tracked from 2 cells collected from 2 independent experiments). Yellow arrows indicate where a KDM5B mRNA (red dots) exited and re-entered the SG
Supplementary information
Supplementary Information
Supplementary Figures 1–4, legends for Supplementary Tables 1 and 2, and legends for Supplementary Movies 1–12.
Rights and permissions
About this article
Cite this article
Moon, S.L., Morisaki, T., Khong, A. et al. Multicolour single-molecule tracking of mRNA interactions with RNP granules. Nat Cell Biol 21, 162–168 (2019). https://doi.org/10.1038/s41556-018-0263-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-018-0263-4
This article is cited by
-
Optogenetic control of mRNA condensation reveals an intimate link between condensate material properties and functions
Nature Communications (2024)
-
Technologies for studying phase-separated biomolecular condensates
Advanced Biotechnology (2024)
-
Spectroscopic single-molecule localization microscopy: applications and prospective
Nano Convergence (2023)
-
Profiling stress-triggered RNA condensation with photocatalytic proximity labeling
Nature Communications (2023)
-
A guide to membraneless organelles and their various roles in gene regulation
Nature Reviews Molecular Cell Biology (2023)