Abstract
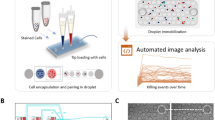
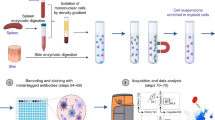
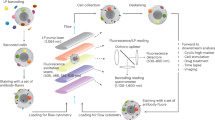
Assays for assessing cell-mediated cytotoxicity are largely target-cell-centric and cannot identify and isolate subpopulations of cytotoxic effector cells. Here we describe an assay compatible with flow cytometry for the accurate identification and sorting of functional killer-cell subpopulations in co-cultures. The assay, which we named PAINTKiller (for ‘proximity affinity intracellular transfer identification of killer cells’), relies on the detection of an intracellular fluorescent protein ‘painted’ by a lysed cell on the surface of the lysing cytotoxic cell (specifically, on cell lysis the intracellular fluorescein derivative carboxyfluorescein succinimidyl ester is captured on the surface of the natural killer cell by an antibody for anti-fluorescein isothiocyanate linked to an antibody for the pan-leucocyte surface receptor CD45). The assay can be integrated with single-cell RNA sequencing for the analysis of molecular pathways associated with cell cytotoxicity and may be used to uncover correlates of functional immune responses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Curated hallmark gene sets are publicly available from Molecular Signatures Database (MSigDB, v.7.4). Sequencing data from PAINTKiller-seq are available from the GEO database, with accession number GSE207508. All data needed to evaluate the conclusions described in this paper are included within the paper and its Supplementary Information. The raw and analysed datasets generated during the study are available for research purposes from the corresponding authors on reasonable request. Source data are provided with this paper.
Code availability
The R code to reproduce the analyses shown in the figures is available on Zenodo at https://doi.org/10.5281/zenodo.8004120.
References
Kagi, D., Ledermann, B., Burki, K., Zinkernagel, R. M. & Hengartner, H. Molecular mechanisms of lymphocyte-mediated cytotoxicity and their role in immunological protection and pathogenesis in vivo. Annu. Rev. Immunol. 14, 207–232 (1996).
Prager, I. & Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 105, 1319–1329 (2019).
Brunner, K. T., Mauel, J., Cerottini, J. C. & Chapuis, B. Quantitative assay of the lytic action of immune lymphoid cells on 51Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology 14, 181–196 (1968).
Sepp, A., Binns, R. M. & Lechler, R. I. Improved protocol for colorimetric detection of complement-mediated cytotoxicity based on the measurement of cytoplasmic lactate dehydrogenase activity. J. Immunol. Methods 196, 175–180 (1996).
Wong, P., Wagner, J. A., Berrien-Elliott, M. M., Schappe, T. & Fehniger, T. A. Flow cytometry-based ex vivo murine NK cell cytotoxicity assay. STAR Protoc. 2, 100262 (2021).
Kandarian, F., Sunga, G. M., Arango-Saenz, D. & Rossetti, M. A flow cytometry-based cytotoxicity assay for the assessment of human NK cell activity. J. Vis. Exp. (126), e56191 (2017).
Wu, X., Zhang, Y., Li, Y. & Schmidt-Wolf, I. G. H. Improvements in flow cytometry-based cytotoxicity assay. Cytometry A 99, 680–688 (2021).
Peper, J. K. et al. An impedance-based cytotoxicity assay for real-time and label-free assessment of T-cell-mediated killing of adherent cells. J. Immunol. Methods 405, 192–198 (2014).
Vembadi, A., Menachery, A. & Qasaimeh, M. A. Cell cytometry: review and perspective on biotechnological advances. Front. Bioeng. Biotechnol. 7, 147 (2019).
Slota, M., Lim, J. B., Dang, Y. & Disis, M. L. ELISpot for measuring human immune responses to vaccines. Expert Rev. Vaccines 10, 299–306 (2011).
Alter, G., Malenfant, J. M. & Altfeld, M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods 294, 15–22 (2004).
Bryceson, Y. T., March, M. E., Barber, D. F., Ljunggren, H. G. & Long, E. O. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J. Exp. Med. 202, 1001–1012 (2005).
Wolint, P., Betts, M. R., Koup, R. A. & Oxenius, A. Immediate cytotoxicity but not degranulation distinguishes effector and memory subsets of CD8+ T cells. J. Exp. Med. 199, 925–936 (2004).
Skelley, A. M., Kirak, O., Suh, H., Jaenisch, R. & Voldman, J. Microfluidic control of cell pairing and fusion. Nat. Methods 6, 147–152 (2009).
Dura, B. et al. Profiling lymphocyte interactions at the single-cell level by microfluidic cell pairing. Nat. Commun. 6, 5940 (2015).
Dura, B. et al. Longitudinal multiparameter assay of lymphocyte interactions from onset by microfluidic cell pairing and culture. Proc. Natl Acad. Sci. USA 113, E3599–E3608 (2016).
Assenmacher, M., Lohning, M. & Radbruch, A. Detection and isolation of cytokine secreting cells using the cytometric cytokine secretion assay. Curr. Protoc. Immunol. https://doi.org/10.1002/0471142735.im0627s46 (2002).
McKinnon, K. M. Flow cytometry: an overview. Curr. Protoc. Immunol. https://doi.org/10.1002/cpim.40 (2018).
Deng, N. & Mosmann, T. R. Optimization of the cytokine secretion assay for human IL-2 in single and combination assays. Cytometry A 87, 777–783 (2015).
Trinklein, N. D. et al. Efficient tumor killing and minimal cytokine release with novel T-cell agonist bispecific antibodies. MAbs 11, 639–652 (2019).
Good, Z. et al. Proliferation tracing with single-cell mass cytometry optimizes generation of stem cell memory-like T cells. Nat. Biotechnol. 37, 259–266 (2019).
Matera, G., Lupi, M. & Ubezio, P. Heterogeneous cell response to topotecan in a CFSE-based proliferation test. Cytometry A 62, 118–128 (2004).
Ponchio, L. et al. Mitomycin C as an alternative to irradiation to inhibit the feeder layer growth in long-term culture assays. Cytotherapy 2, 281–286 (2000).
Llames, S., Garcia-Perez, E., Meana, A., Larcher, F. & del Rio, M. Feeder layer cell actions and applications. Tissue Eng. B Rev. 21, 345–353 (2015).
Krzewski, K., Gil-Krzewska, A., Nguyen, V., Peruzzi, G. & Coligan, J. E. LAMP1/CD107a is required for efficient perforin delivery to lytic granules and NK-cell cytotoxicity. Blood 121, 4672–4683 (2013).
Liu, D. et al. Integrin-dependent organization and bidirectional vesicular traffic at cytotoxic immune synapses. Immunity 31, 99–109 (2009).
Suen, W. C., Lee, W. Y., Leung, K. T., Pan, X. H. & Li, G. Natural killer cell-based cancer immunotherapy: a review on 10 years completed clinical trials. Cancer Invest. 36, 431–457 (2018).
Surman, D. R., Dudley, M. E., Overwijk, W. W. & Restifo, N. P. Cutting edge: CD4+ T cell control of CD8+ T cell reactivity to a model tumor antigen. J. Immunol. 164, 562–565 (2000).
Rosenberg, S. A. & Dudley, M. E. Cancer regression in patients with metastatic melanoma after the transfer of autologous antitumor lymphocytes. Proc. Natl Acad. Sci. USA 101, 14639–14645 (2004).
Caligiuri, M. A. Human natural killer cells. Blood 112, 461–469 (2008).
Spiegel, J. Y. et al. CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: a phase 1 trial. Nat. Med. 27, 1419–1431 (2021).
Prager, I. et al. NK cells switch from granzyme B to death receptor-mediated cytotoxicity during serial killing. J. Exp. Med. 216, 2113–2127 (2019).
Park, J. et al. Multifunctional microparticles with stimulation and sensing capabilities for facile NK cell activity assay. ACS Sens. 6, 693–697 (2021).
Dybkaer, K. et al. Genome wide transcriptional analysis of resting and IL2 activated human natural killer cells: gene expression signatures indicative of novel molecular signaling pathways. BMC Genomics 8, 230 (2007).
Zhang, J. et al. Sequential actions of EOMES and T-BET promote stepwise maturation of natural killer cells. Nat. Commun. 12, 5446 (2021).
van Helden, M. J. et al. Terminal NK cell maturation is controlled by concerted actions of T-bet and Zeb2 and is essential for melanoma rejection. J. Exp. Med. 212, 2015–2025 (2015).
Chester, C., Fritsch, K. & Kohrt, H. E. Natural killer cell immunomodulation: targeting activating, inhibitory, and co-stimulatory receptor signaling for cancer immunotherapy. Front. Immunol. 6, 601 (2015).
Connor, J. H., Weiser, D. C., Li, S., Hallenbeck, J. M. & Shenolikar, S. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell. Biol. 21, 6841–6850 (2001).
Wang, X. W. et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl Acad. Sci. USA 96, 3706–3711 (1999).
Li, Y., Yu, M., Yin, J., Yan, H. & Wang, X. Enhanced calcium signal induces NK cell degranulation but inhibits its cytotoxic activity. J. Immunol. 208, 347–357 (2022).
Jacobi, C. et al. Exposure of NK cells to intravenous immunoglobulin induces IFN gamma release and degranulation but inhibits their cytotoxic activity. Clin. Immunol. 133, 393–401 (2009).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Parish, C. R. Fluorescent dyes for lymphocyte migration and proliferation studies. Immunol. Cell Biol. 77, 499–508 (1999).
Zheng, Y., Tang, L., Mabardi, L., Kumari, S. & Irvine, D. J. Enhancing adoptive cell therapy of cancer through targeted delivery of small-molecule immunomodulators to internalizing or noninternalizing receptors. ACS Nano 11, 3089–3100 (2017).
Wu, T., Womersley, H. J., Wang, J. R., Scolnick, J. & Cheow, L. F. Time-resolved assessment of single-cell protein secretion by sequencing. Nat. Methods 20, 723–734 (2023).
Roelli, P., bbimber, Flynn, B., santiagorevale & Gui, G. Hoohm/CITE-seq-Count: 1.4.2 (1.4.2). Zenodo https://doi.org/10.5281/zenodo.2585469 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Pont, F., Tosolini, M. & Fournie, J. J. Single-Cell Signature Explorer for comprehensive visualization of single cell signatures across scRNA-seq datasets. Nucleic Acids Res. 47, e133 (2019).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Acknowledgements
We acknowledge technical support from the Flow Cytometry Laboratory (NUS Medical Sciences Cluster) for cell sorting. L.F.C. was supported for the research described in this study by the National Medical Research Council (MOH-OFIRG18nov-003), the Ministry of Education of Singapore (MOE-T2EP30120-0008) and Singapore-MIT Alliance for Research and Technology Critical Analytics for Manufacturing of Personalized-Medicine (SMART CAMP) Interdisciplinary Research Group.
Author information
Authors and Affiliations
Contributions
L.F.C., Y.H.L. and T.W. conceived and designed the study. T.W. performed the experiments corresponding to Figs. 2, 4 and 7. Y.H.L. performed the experiments corresponding to Fig. 3. T.W. and L.F.C. performed the PAINTKiller-seq data analysis. L.F.C., T.W. and Y.H.L. wrote the manuscript. All authors commented on the manuscript and approved the submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Sample demultiplexing of PAINTKiller-seq.
a, Unfiltered gene-expression data from two sequencing lanes (‘Exp. 1’ and ‘Exp. 2’). Cells with the following criteria were filtered for downstream analysis: gene numbers > 1,200 and < 8,000, gene-expression counts > 1,000 and < 40,000, mitochondrial genes < 10% of total genes detected. b, The cell samples, including NK-K562 co-culture group (‘NK + K562’), NK only group (‘NK only’) and isotype staining control (‘Isotype ctrl’), were demultiplexed by their staining intensity of anti-β2M hashtags. Cell numbers for each group were shown in bracket.
Extended Data Fig. 2 NK cell identification and cell clustering.
a, b, Cells were clustered by their transcriptional profiles (a), and non-NK cells were excluded according to the expression of lineage-associated genes such as CD3E for T cells and HBA1/HBA2 for K562 cells (b). c, NK cells were re-grouped by their transcriptional profiles. A total of 13 clusters were identified. Cluster 6 and 7 were mostly found in NK sample that has been co-cultured with K562 cells.
Supplementary information
Source data
Source Data for Fig. 2
Source data.
Source Data for Fig. 3
Source data.
Source Data for Fig. 4
Source data.
Source Data for Fig. 5
Source data.
Source Data for Fig. 6
Source data.
Source Data for Fig. 7
Source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luah, Y.H., Wu, T. & Cheow, L.F. Identification, sorting and profiling of functional killer cells via the capture of fluorescent target-cell lysate. Nat. Biomed. Eng 8, 248–262 (2024). https://doi.org/10.1038/s41551-023-01089-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-023-01089-z