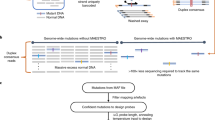
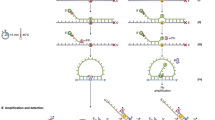
Abstract
The efficiency of DNA-enrichment techniques is often insufficient to detect mutations that occur at low frequencies. Here we report a DNA-excision method for the detection of low-frequency mutations in genomic DNA and in circulating cell-free DNA at single-nucleotide resolution. The method is based on a competitive DNA-binding-and-digestion mechanism, effected by deoxyribonuclease I (DNase) guided by single-stranded phosphorothioated DNA (sgDNase), for the removal of wild-type DNA strands. The sgDNase can be designed against any wild-type DNA sequences, allowing for the uniform enrichment of all the mutations within the target-binding region of single-stranded phosphorothioated DNA at mild-temperature conditions. Pretreatment with sgDNase enriches all mutant strands with initial frequencies down to 0.01% and leads to high discrimination factors for all types of single-nucleotide mismatch in multiple sequence contexts, as we show for the identification of low-abundance mutations in samples of blood or tissue from patients with cancer. The method can be coupled with next-generation sequencing, droplet digital polymerase chain reaction, Sanger sequencing, fluorescent-probe-based assays and other mutation-detection methods.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The main data supporting the results in this study are available within the paper and its Supplementary Information. All data generated during the study are available from figshare with the identifier https://doi.org/10.6084/m9.figshare.23529204. Source data for the figures are provided with this paper.
References
Vogelstein, B. et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525–532 (1988).
Abascal, F. et al. Somatic mutation landscapes at single-molecule resolution. Nature 593, 405–410 (2021).
Diaz, L. A. & Bardelli, A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 32, 579–586 (2014).
Abbosh, C., Birkbak, N. J. & Swanton, C. Early stage NSCLC—challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 15, 577–586 (2018).
Lo, Y. M. D., Han, D. S. C., Jiang, P. & Chiu, R. W. K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 372, eaaw3616 (2021).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
Baker, M. Digital PCR hits its stride. Nat. Methods 9, 541–544 (2012).
Bielas, J. H. & Loeb, L. A. Quantification of random genomic mutations. Nat. Methods 2, 285–290 (2005).
Dressman, D., Yan, H., Traverso, G., Kinzler, K. W. & Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA 100, 8817–8822 (2003).
Das, J., Ivanov, I., Sargent, E. H. & Kelley, S. O. DNA clutch probes for circulating tumor DNA analysis. J. Am. Chem. Soc. 138, 11009–11016 (2016).
Kutyavin, I. V. et al. 3′-minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28, 655–661 (2000).
Singh, S. K., Nielsen, P., Koshkin, A. A. & Wengel, J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 455–456 (1998).
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012).
Milbury, C. A., Li, J. & Makrigiorgos, G. M. PCR-based methods for the enrichment of minority alleles and mutations. Clin. Chem. 55, 632–640 (2009).
Li, J. et al. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 14, 579–584 (2008).
How Kit, A. et al. Sensitive detection of KRAS mutations using enhanced-ice-COLD-PCR mutation enrichment and direct sequence identification. Hum. Mutat. 34, 1568–1580 (2013).
Dominguez, P. L. & Kolodney, M. S. Wild-type blocking polymerase chain reaction for detection of single nucleotide minority mutations from clinical specimens. Oncogene 24, 6830–6834 (2005).
Oldenburg, R. P., Liu, M. S. & Kolodney, M. S. Selective amplification of rare mutations using locked nucleic acid oligonucleotides that competitively inhibit primer binding to wild-type DNA. J. Invest. Dermatol. 128, 398–402 (2008).
Wu, L. R., Chen, S. X., Wu, Y., Patel, A. A. & Zhang, D. Y. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat. Biomed. Eng. 1, 714–723 (2017).
Song, P. et al. Selective multiplexed enrichment for the detection and quantitation of low-fraction DNA variants via low-depth sequencing. Nat. Biomed. Eng. 5, 690–701 (2021).
Darbeheshti, F., Yu, F., Ahmed, F., Adalsteinsson, V. A. & Makrigiorgos, G. M. Recent developments in mutation enrichment and detection technologies. Clin. Chem. 68, 1250–1260 (2022).
Song, C. et al. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res. 44, e146 (2016).
Zhulidov, P. A. et al. Simple cDNA normalization using kamchatka crab duplex-specific nuclease. Nucleic Acids Res. 32, e37 (2004).
Gu, W. et al. Depletion of abundant sequences by hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 17, 41 (2016).
Fu, Y. et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31, 822–826 (2013).
Song, J. et al. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 48, e19 (2020).
Liu, Q. et al. Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations. Nucleic Acids Res. 49, e75 (2021).
Haldar, T. et al. Unexpected complexity in the products arising from NaOH-, heat-, amine-, and glycosylase-induced strand cleavage at an abasic site in DNA. Chem. Res. Toxicol. 35, 218–232 (2022).
Xiao, X., Wu, T., Gu, F. & Zhao, M. Generation of artificial sequence-specific nucleases via a preassembled inert-template. Chem. Sci. 7, 2051–2057 (2016).
Paez, J. G. et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304, 1497–1500 (2004).
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Gilar, M. et al. Ion-pair reversed-phase high-performance liquid chromatography analysis of oligonucleotides: retention prediction. J. Chromatogr. A 958, 167–182 (2002).
Wu, T. et al. DNA terminal structure-mediated enzymatic reaction for ultra-sensitive discrimination of single nucleotide variations in circulating cell-free DNA. Nucleic Acids Res. 46, e24 (2018).
Shepherd, F. A. et al. Erlotinib in previously treated non-small-cell lung cancer. N. Engl. J. Med. 353, 123–132 (2005).
Soria, J. C. et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N. Engl. J. Med. 378, 113–125 (2018).
Hoshika, S. et al. Hachimoji DNA and RNA: a genetic system with eight building blocks. Science 363, 884–887 (2019).
Watkins, N. E. Jr. & SantaLucia, J. Jr. Nearest-neighbor thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res. 33, 6258–6267 (2005).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011).
Limaye, N. et al. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat. Genet. 41, 118–124 (2009).
Soblet, J. et al. Blue Rubber Bleb Nevus (BRBN) syndrome is caused by somatic TEK (TIE2) mutations. J. Invest. Dermatol. 137, 207–216 (2017).
Martinez-Lopez, A. et al. CLOVES syndrome: review of a PIK3CA-related overgrowth spectrum (PROS). Clin. Genet. 91, 14–21 (2017).
Wassef, M. et al. Vascular anomalies classification: recommendations from the International Society for the Study of Vascular Anomalies. Pediatrics 136, E203–E214 (2015).
Kurreck, J. Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem. 270, 1628–1644 (2003).
Stein, C. A. & Cheng, Y. C. Antisense oligonucleotides as therapeutic agents—is the bullet really magical. Science 261, 1004–1012 (1993).
Liang, C. & Allen, L. C. Sulfur does not form double bonds in phosphorothioate anions. J. Am. Chem. Soc. 109, 6449–6453 (1987).
Wang, L. et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat. Chem. Biol. 3, 709–710 (2007).
Pechenaya, V. I. Influence of electrostatic interactions on the sugar phosphate backbone conformation in DNA. Biopolymers 33, 37–44 (1993).
Horton, N. C., Connolly, B. A. & Perona, J. J. Inhibition of EcoRV endonuclease by deoxyribo-3′-S-phosphorothiolates: a high-resolution X-ray crystallographic study. J. Am. Chem. Soc. 122, 3314–3324 (2000).
Zhang, Y. C. et al. Theoretical study on steric effects of DNA phosphorothioation: B-helical destabilization in Rp-phosphorothioated DNA. J. Phys. Chem. B 116, 10639–10648 (2012).
Thorogood, H., Grasby, J. A. & Connolly, B. A. Influence of the phosphate backbone on the recognition and hydrolysis of DNA by the EcoRV restriction endonuclease—a study using oligodeoxynucleotide phosphorothioates. J. Biol. Chem. 271, 8855–8862 (1996).
Acknowledgements
We thank the National Center for Protein Science at Peking University (Beijing, China) for assistance with droplet digital PCR. This work was supported by the National Natural Science Foundation of China (22174005, 21974005, 21575008).
Author information
Authors and Affiliations
Contributions
M. Zhao conceived the project. M. Zhao, W.C., T.W. and X.X. performed the sequences and experiments design. W.C., H.X. and S.D. conducted the experiments and data analysis. J.W., Z.Y., Y.J. and M. Zou helped improve some experiments. Z.L., W.Y. and B.Z. provided blood and tissue samples from patients with venous malformations. X.X. provided blood and tissue samples from patients with NSCLC or colorectal cancer. W.C., T.W. and M. Zhao wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Peking University holds an issued Chinese patent (patent number: ZL 2019 1 0052680.4) on this work. The authors declare no other competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Feng Li, David Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Multiplexed sgDNase pretreatment.
Next-generation sequencing results of the five EGFR and KRAS mutations (EGFR L858R, EGFR 19del, EGFR G719S, EGFR T790M and KRAS G13D) in mixed synthetic samples before (a1-a5) and after (b1-b5) 5-plex sgDNase pretreatment. The initial mutation abundance was 0.4% for EGFR G719S and 0.1% for other tested mutation. The five target mutations were pretreated by sgDNase simultaneously. The variant to noise plots derived from Illumina MiSeq sequencing data are depicted.
Extended Data Fig. 2 Detection of low-abundance mutations in clinical samples with sgDNase pretreatment.
NGS measurement results of the abundance of EGFR L858R mutation in a gDNA (P2-T) and b cfDNA (P2-B) without sgDNase pretreatment. Variant to noise plots derived from Illumina MiSeq sequencing data are depicted.
Extended Data Fig. 3 Summary of sample test results with sgDNase pretreatment.
a, Beeswarm plot of post-VAFs of 309 negative control gDNA samples and 267 positive samples with pre-VAF at 0.01% after sgDNase pretreatment. b, Receiver operator characteristic (ROC) plot for post-VAFs of samples in a after pretreatment by sgDNase.
Supplementary information
Supplementary Information
Supplementary methods, discussion, figures, tables and references.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, W., Xu, H., Dai, S. et al. Detection of low-frequency mutations in clinical samples by increasing mutation abundance via the excision of wild-type sequences. Nat. Biomed. Eng 7, 1602–1613 (2023). https://doi.org/10.1038/s41551-023-01072-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-023-01072-8