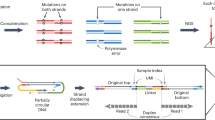
Abstract
Assaying for large numbers of low-frequency mutations requires sequencing at extremely high depth and accuracy. Increasing sequencing depth aids the detection of low-frequency mutations yet limits the number of loci that can be simultaneously probed. Here we report a method for the accurate tracking of thousands of distinct mutations that requires substantially fewer reads per locus than conventional hybrid-capture duplex sequencing. The method, which we named MAESTRO (for minor-allele-enriched sequencing through recognition oligonucleotides), combines massively parallel mutation enrichment with duplex sequencing to track up to 10,000 low-frequency mutations, with up to 100-fold fewer reads per locus. We show that MAESTRO can be used to test for chimaerism by tracking donor-exclusive single-nucleotide polymorphisms in sheared genomic DNA from human cell lines, to validate whole-exome sequencing and whole-genome sequencing for the detection of mutations in breast-tumour samples from 16 patients, and to monitor the patients for minimal residual disease via the analysis of cell-free DNA from liquid biopsies. MAESTRO improves the breadth, depth, accuracy and efficiency of mutation testing by sequencing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$99.00 per year
only $8.25 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data supporting the results in this study are available within the paper and its Supplementary Information. Sequencing data have been deposited into the controlled-access database Data Use Oversight System (DUOS; http://duos.broadinstitute.org) under the accession number DUOS-000135.
Code availability
Custom code for designing MAESTRO probes from somatic variant calls can be found at https://github.com/broadinstitute/MAESTRO-probe_designer.
References
Luquette, L. J., Bohrson, C. L., Sherman, M. A. & Park, P. J. Identification of somatic mutations in single cell DNA-seq using a spatial model of allelic imbalance. Nat. Commun. 10, 3908 (2019).
Ludwig, L. S. et al. Lineage tracing in humans enabled by mitochondrial mutations and single-cell genomics. Cell 176, e22 (2019).
Zahn, L. M. Mapping genotype to phenotype. Science 362, 555.4–556 (2018).
D’Gama, A. M. & Walsh, C. A. Somatic mosaicism and neurodevelopmental disease. Nat. Neurosci. 21, 1504–1514 (2018).
Garcia-Murillas, I. et al. Assessment of molecular relapse detection in early-stage breast cancer. JAMA Oncol. https://doi.org/10.1001/jamaoncol.2019.1838 (2019).
Canick, J. A., Palomaki, G. E., Kloza, E. M., Lambert-Messerlian, G. M. & Haddow, J. E. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenat. Diagn. 33, 667–674 (2013).
Bejar, R. et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J. Clin. Oncol. 32, 2691–2698 (2014).
Snyder, T. M., Khush, K. K., Valantine, H. A. & Quake, S. R. Universal noninvasive detection of solid organ transplant rejection. Proc. Natl Acad. Sci. USA 108, 6229–6234 (2011).
Blauwkamp, T. A. et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol 4, 663–674 (2019).
Boyd, S. D. et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci. Transl. Med. 1, 12ra23 (2009).
Gilbert, J. A. et al. Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018).
Lowe, A., Murray, C., Whitaker, J., Tully, G. & Gill, P. The propensity of individuals to deposit DNA and secondary transfer of low level DNA from individuals to inert surfaces. Forensic Sci. Int. 129, 25–34 (2002).
Schmitt, M. W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl Acad. Sci. USA 109, 14508–14513 (2012).
Song, C. et al. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res. 44, e146 (2016).
Li, J. & Makrigiorgos, G. Mike COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing. Biochem. Soc. Trans. 37, 427–432 (2009).
Wu, L. R., Chen, S. X., Wu, Y., Patel, A. A. & Zhang, D. Y. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat. Biomed. Eng. 1, 714–723 (2017).
Jeffreys, A. J. & May, C. A. DNA enrichment by allele-specific hybridization (DEASH): a novel method for haplotyping and for detecting low-frequency base substitutional variants and recombinant DNA molecules. Genome Res. 13, 2316–2324 (2003).
Gaudet, M., Fara, A.-G., Beritognolo, I. & Sabatti, M. Allele-specific PCR in SNP genotyping. Methods Mol. Biol. 578, 415–424 (2009).
Vargas, D. Y., Marras, S. A. E., Tyagi, S. & Kramer, F. R. Suppression of wild-type amplification by selectivity enhancing agents in PCR assays that utilize superselective primers for the detection of rare somatic mutations. J. Mol. Diagn. 20, 415–427 (2018).
Li, J. et al. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 14, 579–584 (2008).
Li, J., Milbury, C. A., Li, C. & Makrigiorgos, G. M. Two-round coamplification at lower denaturation temperature-PCR (COLD-PCR)-based sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Hum. Mutat. 30, 1583–1590 (2009).
Schmitt, M. W. et al. Sequencing small genomic targets with high efficiency and extreme accuracy. Nat. Methods 12, 423–425 (2015).
Parsons, H. A. et al. Sensitive detection of minimal residual disease in patients treated for early-stage breast cancer. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-19-3005 (2020).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Machin, G. Non-identical monozygotic twins, intermediate twin types, zygosity testing, and the non-random nature of monozygotic twinning: a review. Am. J. Med. Genet. C 151C, 110–127 (2009).
Shimoni, A. & Nagler, A. Non-myeloablative stem cell transplantation (NST): chimerism testing as guidance for immune-therapeutic manipulations. Leukemia 15, 1967–1975 (2001).
Breuer, S. et al. Early recipient chimerism testing in the T- and NK-cell lineages for risk assessment of graft rejection in pediatric patients undergoing allogeneic stem cell transplantation. Leukemia 26, 509–519 (2012).
Tyler, J., Kumer, L., Fisher, C., Casey, H. & Shike, H. Personalized chimerism test that uses selection of short tandem repeat or quantitative PCR depending on patient’s chimerism status. J. Mol. Diagn. 21, 483–490 (2019).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 20, 548–554 (2014).
Lee, H., Park, C., Na, W., Park, K. H. & Shin, S. Precision cell-free DNA extraction for liquid biopsy by integrated microfluidics. npj Precis. Oncol. 4, 3 (2020).
Mauger, F. et al. Comparison of commercially available whole-genome sequencing kits for variant detection in circulating cell-free DNA. Sci. Rep. 10, 6190 (2020).
Liu, D. et al. Multiplex cell-free DNA reference materials for quality control of next-generation sequencing-based in vitro diagnostic tests of colorectal cancer tolerance. J. Cancer 9, 3812–3823 (2018).
Tsao, D. S. et al. A novel high-throughput molecular counting method with single base-pair resolution enables accurate single-gene NIPT. Sci. Rep. 9, 14382 (2019).
Pantel, K. & Alix-Panabières, C. Liquid biopsy and minimal residual disease—latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424 (2019).
Tie, J. et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 8, 346ra92 (2016).
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403 (2017).
Coombes, R. C. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
Wan, J. C. M. et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 12, eaaz8084 (2020).
McDonald, B. R. et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 11, eaax7392 (2019).
Butler, T. M. et al. Circulating tumor DNA dynamics using patient-customized assays are associated with outcome in neoadjuvantly treated breast cancer. Cold Spring Harb. Mol. Case Stud. 5, a003772 (2019).
Magbanua, M. J. M. et al. Circulating tumor DNA in neoadjuvant treated breast cancer reflects response and survival. Oncology https://doi.org/10.1101/2020.02.03.20019760 (2020).
Moding, E. J. et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat. Cancer 1, 176–183 (2020).
Etienne, G. et al. Long-term follow-up of the French Stop Imatinib (STIM1) study in patients with chronic myeloid leukemia. J. Clin. Oncol. 35, 298–305 (2017).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Zviran, A. et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 26, 1114–1124 (2020).
Masuda, N. et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N. Engl. J. Med. 376, 2147–2159 (2017).
von Minckwitz, G. et al. Trastuzumab emtansine for residual invasive HER2-positive breast cancer. N. Engl. J. Med. 380, 617–628 (2019).
Zook, J. M. et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol. 37, 561–566 (2019).
Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). 1000.
Adalsteinsson, V. A. et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 8, 1324 (2017).
Köster, J. & Rahmann, S. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522 (2012).
Acknowledgements
We acknowledge the patients and their families for their contributions to this study. We also thank the generous support from the Gerstner Family Foundation. This project was supported in part by the Bridge Project (J.C.L. and V.A.A.), a partnership between the Koch Institute for Integrative Cancer Research at MIT and the Dana-Farber/Harvard Cancer Center. We also acknowledge support from National Institutes of Health grants R33 CA217652 (G.M.M. and V.A.A.) and R01 CA221874 (G.M.M. and V.A.A.).
Author information
Authors and Affiliations
Contributions
G.G., E.N. and J.H.B. designed the research, analysed the data and wrote the manuscript. T.B. and J.R. analysed the data and created figures for the manuscript. S.C.R., D.S., K.X., R.L., F.Y. and K.W.L. designed experiments and interpreted the results. A.D.C., D.G.S., S.M.T., I.E.K. and H.A.P. contributed clinical samples and the interpretation of results. J.C.L., G.M.M., T.R.G. and V.A.A. designed the research, interpreted the results and wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
A.D.C. has advisory-board roles with Clovis, Dendreon and Bayer, and has received research funding from Bayer. S.M.T. has received research funding to the institution from AstraZeneca, Eli Lilly, Merck, Novartis, Nektar, Pfizer, Genentech, Immunomedics, Exelixis, Bristol-Myers Squibb, Eisai, Nanostring, Cyclacel, Sanofi, Odonate and Seattle Genetics, and has received honorariums for consulting and advisory-board participation from AstraZeneca, Eli Lilly, Merck, Novartis, Nektar, Pfizer, Genentech, Immunomedics, Bristol-Myers Squibb, Eisai, Nanostring, Sanofi, Odonate, Seattle Genetics, Puma, Anthenex, OncoPep, Abbvie, G1 Therapeutics, Silverback Therapeutics and Celldex. I.E.K. has received research funding to the institute from Genentech, Pfizer and Daichii-Sankyo, and has received honorariums for consulting and advisory-board participation from Genentech, Daichii-Sankyo, Macrogenics, Context Therapeutics, Taiho Oncology, Merck, Novartis and Bristol-Myers Squibb. H.A.P. has a paid consultant role for Foundation Medicine. T.R.G. is a paid advisor to GlaxoSmithKline, and is a co-founder and equity holder of Sherlock Biosciences and FORMA Therapeutics. V.A.A. has a patent application filed with the Broad Institute, and is a member of the scientific advisory boards of Bertis and AGCT, which were not involved in this study. The other authors report no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary figures.
Supplementary Data 1
Probe sequences.
Supplementary Data 2
Supplementary tables.
Rights and permissions
About this article
Cite this article
Gydush, G., Nguyen, E., Bae, J.H. et al. Massively parallel enrichment of low-frequency alleles enables duplex sequencing at low depth. Nat. Biomed. Eng 6, 257–266 (2022). https://doi.org/10.1038/s41551-022-00855-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-022-00855-9
This article is cited by
-
An in vitro CRISPR screen of cell-free DNA identifies apoptosis as the primary mediator of cell-free DNA release
Communications Biology (2024)
-
Bridging horizons beyond CIRCULATE-Japan: a new paradigm in molecular residual disease detection via whole genome sequencing-based circulating tumor DNA assay
International Journal of Clinical Oncology (2024)
-
Perspectives for the clinical application of ctDNA analysis to breast cancer drug therapy
Breast Cancer (2024)
-
Liquid biopsy for brain metastases and leptomeningeal disease in patients with breast cancer
npj Breast Cancer (2023)
-
Minimal residual disease in solid tumors: an overview
Frontiers of Medicine (2023)